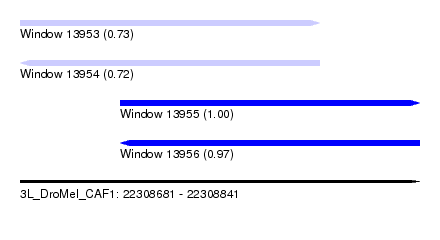
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,308,681 – 22,308,841 |
| Length | 160 |
| Max. P | 0.999873 |

| Location | 22,308,681 – 22,308,801 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -32.91 |
| Energy contribution | -32.91 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22308681 120 + 23771897 UCCAAUCUACUCCCGGCCAUUACGCACACCCACUGGGGCGGGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCA ..............(((((...(((...((....)).)))(((..(((((......))))))))........)))))........(((((((..(....)..)))).)))((.....)). ( -34.10) >DroSec_CAF1 67347 120 + 1 UCCAAUCUACUCCCGGCCAUUACGCACACCCACUGGGGCGGGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCA ..............(((((...(((...((....)).)))(((..(((((......))))))))........)))))........(((((((..(....)..)))).)))((.....)). ( -34.10) >DroSim_CAF1 49389 120 + 1 UCCAAUCUACUCCCGGCCAUUACGCACACCCACUGGGGCGGGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCA ..............(((((...(((...((....)).)))(((..(((((......))))))))........)))))........(((((((..(....)..)))).)))((.....)). ( -34.10) >DroEre_CAF1 65825 120 + 1 UCCAAUCUACGCCCGGCCAUUGCGAACACCCACUGGGGCGGGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCA ...............((..(((((((...((((..((((((((....))))...(((..(.(((((.....))))))..)))...))))..))))...)))))))...))((.....)). ( -37.60) >DroYak_CAF1 43009 120 + 1 UCCCAUCUACUCCCGGCCAUUACGCACACCCACUGGGGCGGGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAUCCAGCUUUUGUGCCAAUUUGCAGACAGCGCUACUUGCA ..............(((((...(((...((....)).)))(((..(((((......))))))))........)))))........((((((((........))))).)))((.....)). ( -33.20) >consensus UCCAAUCUACUCCCGGCCAUUACGCACACCCACUGGGGCGGGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCA ...........(((((................)))))((((((....))))....(((((.(((((.....))))))........((((((((........))))).)))))))...)). (-32.91 = -32.91 + 0.00)


| Location | 22,308,681 – 22,308,801 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -41.98 |
| Consensus MFE | -37.98 |
| Energy contribution | -38.42 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22308681 120 - 23771897 UGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCCCGCCCCAGUGGGUGUGCGUAAUGGCCGGGAGUAGAUUGGA .((.....))(((.((.((..(((((((((...(((((..((.((((((.....)))))).))......((((....))))...)))))...)))).))))).)).)).)))........ ( -41.00) >DroSec_CAF1 67347 120 - 1 UGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCCCGCCCCAGUGGGUGUGCGUAAUGGCCGGGAGUAGAUUGGA .((.....))(((.((.((..(((((((((...(((((..((.((((((.....)))))).))......((((....))))...)))))...)))).))))).)).)).)))........ ( -41.00) >DroSim_CAF1 49389 120 - 1 UGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCCCGCCCCAGUGGGUGUGCGUAAUGGCCGGGAGUAGAUUGGA .((.....))(((.((.((..(((((((((...(((((..((.((((((.....)))))).))......((((....))))...)))))...)))).))))).)).)).)))........ ( -41.00) >DroEre_CAF1 65825 120 - 1 UGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCCCGCCCCAGUGGGUGUUCGCAAUGGCCGGGCGUAGAUUGGA .((.....)).(.((((((.(((((((.(((....((((....((((((.....)))))).))))..))))))))))(((((..(((.((.(....).)).))).))))))))))).).. ( -43.20) >DroYak_CAF1 43009 120 - 1 UGCAAGUAGCGCUGUCUGCAAAUUGGCACAAAAGCUGGAUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCCCGCCCCAGUGGGUGUGCGUAAUGGCCGGGAGUAGAUGGGA .((.....)).((((((((...(..((......))..).(.((((((.......((((....))))((((.(((...((((((....)))))).))).)))))))))).))))))))).. ( -43.70) >consensus UGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCCCGCCCCAGUGGGUGUGCGUAAUGGCCGGGAGUAGAUUGGA .((.....))(((.((.((..(((((((((...(((((..((.((((((.....)))))).))......((((....))))...)))))...)))).))))).)).)).)))........ (-37.98 = -38.42 + 0.44)



| Location | 22,308,721 – 22,308,841 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -27.84 |
| Energy contribution | -27.68 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.33 |
| SVM RNA-class probability | 0.999873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22308721 120 + 23771897 GGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCAUUAAAUUGCAAAUCCACACAAACACAUUAAAACACAAUUA (((..(((((......)))))(((((.....))))).........)))((((((...((((((((.....((.....))......))))))))...)))))).................. ( -27.90) >DroSec_CAF1 67387 120 + 1 GGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCAUCAAAUUGCAAAUACACACAAACACAUUAAAACACAAUUA (((..(((((......)))))(((((.....))))).........)))((((((...((((((((.....((.....))......))))))))...)))))).................. ( -27.90) >DroSim_CAF1 49429 120 + 1 GGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCAUCAAAUUGCAAAUCCACACAAACACAUUAAAACACAAUUA (((..(((((......)))))(((((.....))))).........)))((((((...((((((((.....((.....))......))))))))...)))))).................. ( -27.90) >DroEre_CAF1 65865 120 + 1 GGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCAUCAAAUUGCAAGUCCACACAAACACAUUAAAACACAAUUA (((((((((((((.((((...(((((.....)))))......))))...))).))))))).((.....)))))(((((((......)))))))........................... ( -30.40) >DroYak_CAF1 43049 120 + 1 GGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAUCCAGCUUUUGUGCCAAUUUGCAGACAGCGCUACUUGCAUCAAAUUGCAAAUCCACACAAACACAUUAAAACACAAUUA (((..(((((......)))))(((((.....))))).........)))((((((...((((((((.....((.....))......))))))))...)))))).................. ( -26.80) >consensus GGCAAUUGCCCAAUUUGGCGAGCCAAUUAUUUUGGCCAACAACCAGCUUUUGUGGCAAUUUGCAGACAGCGCUACUUGCAUCAAAUUGCAAAUCCACACAAACACAUUAAAACACAAUUA (((..(((((......)))))(((((.....))))).........)))((((((...((((((((.....((.....))......))))))))...)))))).................. (-27.84 = -27.68 + -0.16)


| Location | 22,308,721 – 22,308,841 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -31.74 |
| Energy contribution | -31.78 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22308721 120 - 23771897 UAAUUGUGUUUUAAUGUGUUUGUGUGGAUUUGCAAUUUAAUGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCC (((((((.(......(((((((((..((((((((...((.(((.....))).))..))))))))..))))))(((..((((((......))))))..))))))......).))))))).. ( -33.40) >DroSec_CAF1 67387 120 - 1 UAAUUGUGUUUUAAUGUGUUUGUGUGUAUUUGCAAUUUGAUGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCC .....................((((.((((((((......)))))))))))).....((((.(((((.(((....((((....((((((.....)))))).))))..)))))))))))). ( -33.10) >DroSim_CAF1 49429 120 - 1 UAAUUGUGUUUUAAUGUGUUUGUGUGGAUUUGCAAUUUGAUGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCC (((((((.(......(((((((((..((((((((....(.(((.....))))....))))))))..))))))(((..((((((......))))))..))))))......).))))))).. ( -33.50) >DroEre_CAF1 65865 120 - 1 UAAUUGUGUUUUAAUGUGUUUGUGUGGACUUGCAAUUUGAUGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCC .....................((((..(((((((......))))))).)))).....((((.(((((.(((....((((....((((((.....)))))).))))..)))))))))))). ( -35.40) >DroYak_CAF1 43049 120 - 1 UAAUUGUGUUUUAAUGUGUUUGUGUGGAUUUGCAAUUUGAUGCAAGUAGCGCUGUCUGCAAAUUGGCACAAAAGCUGGAUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCC (((((((........((.(((((((.((((((((....(.(((.....))))....)))))))).))))))).))(((.((..((((((.....)))))))))))......))))))).. ( -35.60) >consensus UAAUUGUGUUUUAAUGUGUUUGUGUGGAUUUGCAAUUUGAUGCAAGUAGCGCUGUCUGCAAAUUGCCACAAAAGCUGGUUGUUGGCCAAAAUAAUUGGCUCGCCAAAUUGGGCAAUUGCC ...(((((...((((.(((..((((..(((((((......))))))).)))).....))).)))).))))).(((..((((((......))))))..))).(((......)))....... (-31.74 = -31.78 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:54 2006