
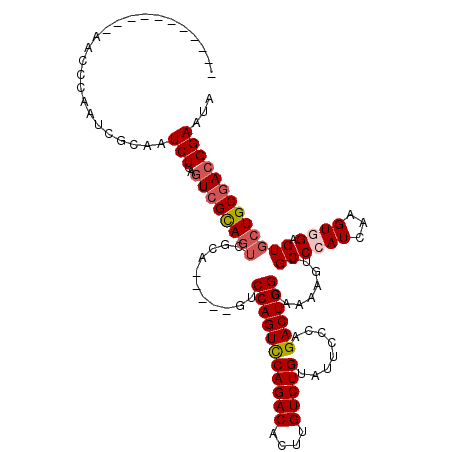
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,307,842 – 22,308,047 |
| Length | 205 |
| Max. P | 0.825433 |

| Location | 22,307,842 – 22,307,943 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.64 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22307842 101 - 23771897 -------------------UGGGACUGGGACCAGGUCCUAGGCCUUAUUCCUUUGAUCUUUUGUCUUUUGCAAUUACAAUUAUGUUUUAUUUGCUCUAGUUGACACAAGAGACAAAAGUU -------------------.(((.(((((((...))))))).)))............((((((((((((((((((((((.((.....)).)))...))))))...))))))))))))).. ( -29.40) >DroSec_CAF1 66474 99 - 1 -------------------UGGGACUGGGACCAGGUCCUAGGCCAUAUUCCUUUGAUCUUUUGUCUUUUGCAAUUACAAUUAUGUUUUAUUUGCUCUAGUUGACACAAGAGGCGAAGA-- -------------------(((..(((((((...))))))).))).....(((((.((((.((((....((((..(((....))).....)))).......)))).))))..))))).-- ( -27.40) >DroSim_CAF1 48525 101 - 1 -------------------UGGGACUGGGACCAGGUCCUAGGCCAUAUUCCUUUGAUCUUUUGUCUUUUGCAAUUACAAUUAUGUUUUAUUUGCUCUAGUUGACACAAGAGACGAAAGUU -------------------(((..(((((((...))))))).)))............((((((((((((((((((((((.((.....)).)))...))))))...))))))))))))).. ( -29.00) >DroEre_CAF1 64932 118 - 1 UUGGGAUUGGGAUGGGGAUUGGGCUUGGGACCAGGUCCUAGGACAUUUUCCUUUGAUCUUCUGUCUCUCGCAACUACAAUUA--UUUUAUUUGCUCUAGUUGACACAAGAAACGAAAGUU (((.....(((((((((((((((((((....))))))).((((.....))))..))))))).)))))..((((((((((...--......)))...)))))).).)))..(((....))) ( -30.90) >DroYak_CAF1 42176 114 - 1 UUGGGUUUGCGUU------UGGUAUUGGGGCCAGGGCCUAGAAAAUUUUUCUUUGAUCUUUUGUCUUUUGCAAUUACAAUGAGGUUUUAUUUGCUCUAGUUGACACAAGAGAGGAAGGUU (((((((.....(------((((......))))))))))))..((((((((((...((((.((((....((((.((.(((...))).)).)))).......)))).)))))))))))))) ( -23.70) >consensus ___________________UGGGACUGGGACCAGGUCCUAGGCCAUAUUCCUUUGAUCUUUUGUCUUUUGCAAUUACAAUUAUGUUUUAUUUGCUCUAGUUGACACAAGAGACGAAAGUU .....................((.(((((((...))))))).)).............((((((((((((((((((((((.((.....)).)))...))))))...))))))))))))).. (-21.20 = -21.64 + 0.44)



| Location | 22,307,943 – 22,308,047 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.26 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22307943 104 + 23771897 -----------AACCCAAUCACAAUCUGAGUCGCAGUCGAA-----GUCCAGUUCAGACACAUGUCUGUAUUCCCAGACUGGCAAAAGUUGCCCAUCAAGUGGAGUGCUGCGACGGAAUA -----------...((..(((.....)))((((((((...(-----.((((.(((((((....)))))........((..((((.....))))..)))).)))).))))))))))).... ( -33.10) >DroSec_CAF1 66573 104 + 1 -----------AACCCAAUCGCAAUCUCAGUCGCAGUCGCA-----GUCCAGUCCAGACACUUGUCUGUAUUCCCAGACUGGAAAAAGUUGCCCAUCAAGUGGAGUGCUGCGACGGAAUA -----------...............((.(((((((.(((.-----.((((((((((((....)))))........))))))).........((((...)))).)))))))))).))... ( -35.10) >DroSim_CAF1 48626 104 + 1 -----------AACCCAAUCGCAAUCUCAUUCGUAGUCGCA-----GUCCAGUCCAGACACUUGUCUGUAUUCCCAGACUGGCAAAAGUUGCCCAUCAAGUGGAGUGCUGCGACGGAAUA -----------.................((((...((((((-----(((((((((((((....)))))........))))))........((((((...)))).)))))))))).)))). ( -31.50) >DroEre_CAF1 65050 120 + 1 ACACAAGCCCAAAAGCAAUCGCAAUCUCAGUCGCAGUCGCAGUCCAGUCCAGUCCAGACACUUGUCUGUAUUCCCAGACUGGCAAAAGUUGCCCAUCAAGUCGAGUGCUGCGACGGAAUA ........((....((....)).......(((((((.(((.(.((((((.(((.(((((....))))).)))....)))))))............((.....)))))))))))))).... ( -35.00) >DroYak_CAF1 42290 102 + 1 ACGCAAACC------CAAUCGCAAUCUUAGUCGCA-----------GCCCAGUCCAGACACUUGUCUGUAUUCCCAGACUGGCAAAAGUUGCCCAUCAAGUCGAGUGCUGC-ACGGAAUA ..((.....------.....)).......((.(((-----------((.(....(((((....)))))........((((((((.....)))).....))))..).)))))-))...... ( -24.10) >consensus ___________AACCCAAUCGCAAUCUCAGUCGCAGUCGCA_____GUCCAGUCCAGACACUUGUCUGUAUUCCCAGACUGGCAAAAGUUGCCCAUCAAGUGGAGUGCUGCGACGGAAUA ........................(((..((((((((...........(((((((((((....)))))........))))))........((((((...)))).)))))))))))))... (-23.94 = -24.82 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:49 2006