
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,307,019 – 22,307,169 |
| Length | 150 |
| Max. P | 0.999772 |

| Location | 22,307,019 – 22,307,129 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.18 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -23.11 |
| Energy contribution | -23.75 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22307019 110 + 23771897 ACAUUU-----GCCCAUAUGUAUGUG-----CUGUCCAAAUGGCAUGCGAUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAUAUGGUACAGAAAAGUG ...(((-----(.((((((...((((-----((((....))))).(((((((((((.(((((((..((....)).))))))).))))))).)))))))....))))))..))))...... ( -31.80) >DroSec_CAF1 65658 115 + 1 ACAUUU-----GCCCAUAUGUAUGUGCUGUGCUGUCCAAAUGGCAUGCGAUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAAAUGGUACAGAAAAGUG ((((..-----......)))).((((((((....(((.....((((((...(((((.(((((((..((....)).))))))).)))))))))))...)))....))))))))........ ( -34.10) >DroSim_CAF1 47693 115 + 1 ACAUUU-----GCCCAUAUGUAUGUGCUGUGCUGUCCAAAUGGCAUGCGAUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAAAUGGUACAGAAAAGUG ((((..-----......)))).((((((((....(((.....((((((...(((((.(((((((..((....)).))))))).)))))))))))...)))....))))))))........ ( -34.10) >DroEre_CAF1 64100 110 + 1 ACAUUU-----GCCCAUAUGUAUGUG-----CCGUCCAAAUGGCAAGCGCUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACUGAAAAAAUGGUUGAGAAAAGUG .(((((-----...(((((((...((-----((((....))))))...)).(((((.(((((((..((....)).))))))).))))))))))(((((........)))))....))))) ( -29.00) >DroYak_CAF1 41257 115 + 1 AUGUAUGUACUGUACAUACAUAUGUG-----AAGUUCAAAUGGCAUGCGAUUUUGCAGCAACAAAGUGGUUGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAAAUGGUACAGGAAAGUG (((((((((...))))))))).((((-----...(((.....((((((...(((((.(((((((..((....)).))))))).)))))))))))....))).......))))........ ( -31.90) >consensus ACAUUU_____GCCCAUAUGUAUGUG_____CUGUCCAAAUGGCAUGCGAUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAAAUGGUACAGAAAAGUG ..............(((.....((((........(((.....((((((...(((((.(((((((..((....)).))))))).)))))))))))...)))........)))).....))) (-23.11 = -23.75 + 0.64)



| Location | 22,307,049 – 22,307,169 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -29.22 |
| Energy contribution | -29.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22307049 120 + 23771897 UGGCAUGCGAUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAUAUGGUACAGAAAAGUGCGCACUGCAAGAAAAAUGGCUAGCUCGGUAGUGCGGGAGA ..((((((...(((((.(((((((..((....)).))))))).))))))))))).............((((......))))(((((((..((............)).)))))))...... ( -34.70) >DroSec_CAF1 65693 120 + 1 UGGCAUGCGAUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAAAUGGUACAGAAAAGUGCGCACUGCAAGAAAAAUGGCUGGCUCAAAAGUGCGGGAAA ..((((((...(((((.(((((((..((....)).))))))).))))))))))).............((((......))))(((((((.((........)).)).....)))))...... ( -32.30) >DroSim_CAF1 47728 120 + 1 UGGCAUGCGAUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAAAUGGUACAGAAAAGUGCGCACUGCAAGAAAAAUGGCUAGCUCAAAAGUGCGGGAAA ..((((((...(((((.(((((((..((....)).))))))).))))))))))).............((((......))))(((((((.((........)).)).....)))))...... ( -32.30) >DroEre_CAF1 64130 120 + 1 UGGCAAGCGCUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACUGAAAAAAUGGUUGAGAAAAGUGCGCACUGAAAGAAAAAUGGCUAACCUAAAAGUGCGGGAAA ......((((((((((.(((((((..((....)).))))))).))........(((((........)))))..))))))))(((((...((.............))...)))))...... ( -31.12) >DroYak_CAF1 41292 120 + 1 UGGCAUGCGAUUUUGCAGCAACAAAGUGGUUGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAAAUGGUACAGGAAAGUGCGCACUGUAAGAAAAAUGGCUAACUUAAAAGUGCGGAAAA ..((((((...(((((.(((((((..((....)).))))))).))))))))))).............((((......))))(((((.((((............))))..)))))...... ( -32.20) >consensus UGGCAUGCGAUUUUGCAGCAACAAAGUGGUAGCAAUUGUUGCGGCAAAGUAUGCAACGGAAAAAAUGGUACAGAAAAGUGCGCACUGCAAGAAAAAUGGCUAGCUCAAAAGUGCGGGAAA ..((((((...(((((.(((((((..((....)).))))))).))))))))))).............((((......))))(((((((.((........)).)).....)))))...... (-29.22 = -29.78 + 0.56)

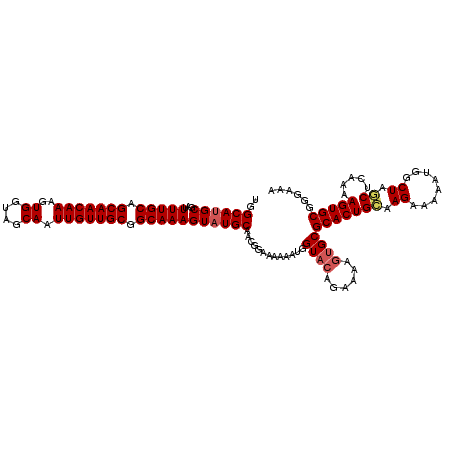

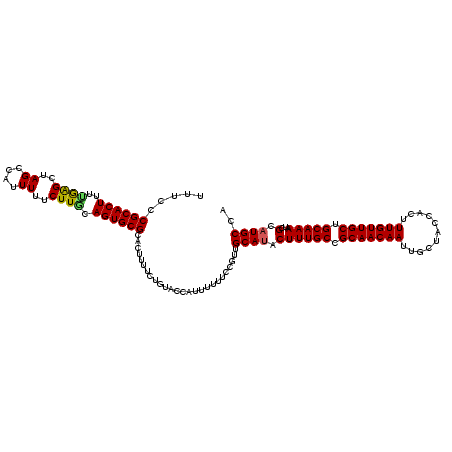
| Location | 22,307,049 – 22,307,169 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.22 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22307049 120 - 23771897 UCUCCCGCACUACCGAGCUAGCCAUUUUUCUUGCAGUGCGCACUUUUCUGUACCAUAUUUUCCGUUGCAUACUUUGCCGCAACAAUUGCUACCACUUUGUUGCUGCAAAAUCGCAUGCCA .....((((((..((((..((....))..)))).))))))..........................((((.((((((.(((((((...........))))))).)))))...).)))).. ( -24.40) >DroSec_CAF1 65693 120 - 1 UUUCCCGCACUUUUGAGCCAGCCAUUUUUCUUGCAGUGCGCACUUUUCUGUACCAUUUUUUCCGUUGCAUACUUUGCCGCAACAAUUGCUACCACUUUGUUGCUGCAAAAUCGCAUGCCA .....((((((.....((.((........)).))))))))..........................((((.((((((.(((((((...........))))))).)))))...).)))).. ( -23.30) >DroSim_CAF1 47728 120 - 1 UUUCCCGCACUUUUGAGCUAGCCAUUUUUCUUGCAGUGCGCACUUUUCUGUACCAUUUUUUCCGUUGCAUACUUUGCCGCAACAAUUGCUACCACUUUGUUGCUGCAAAAUCGCAUGCCA .....((((((.....((.((........)).))))))))..........................((((.((((((.(((((((...........))))))).)))))...).)))).. ( -23.30) >DroEre_CAF1 64130 120 - 1 UUUCCCGCACUUUUAGGUUAGCCAUUUUUCUUUCAGUGCGCACUUUUCUCAACCAUUUUUUCAGUUGCAUACUUUGCCGCAACAAUUGCUACCACUUUGUUGCUGCAAAAGCGCUUGCCA ......(((......((....))..............((((........((((..........)))).....(((((.(((((((...........))))))).))))).)))).))).. ( -25.30) >DroYak_CAF1 41292 120 - 1 UUUUCCGCACUUUUAAGUUAGCCAUUUUUCUUACAGUGCGCACUUUCCUGUACCAUUUUUUCCGUUGCAUACUUUGCCGCAACAAUUGCAACCACUUUGUUGCUGCAAAAUCGCAUGCCA .....((((((..((((..((....))..)))).))))))..........................((((.((((((.(((((((...........))))))).)))))...).)))).. ( -24.00) >consensus UUUCCCGCACUUUUGAGCUAGCCAUUUUUCUUGCAGUGCGCACUUUUCUGUACCAUUUUUUCCGUUGCAUACUUUGCCGCAACAAUUGCUACCACUUUGUUGCUGCAAAAUCGCAUGCCA .....((((((..((((..((....))..)))).))))))..........................((((.((((((.(((((((...........))))))).)))))...).)))).. (-22.46 = -22.22 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:47 2006