
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,305,199 – 22,305,473 |
| Length | 274 |
| Max. P | 0.986426 |

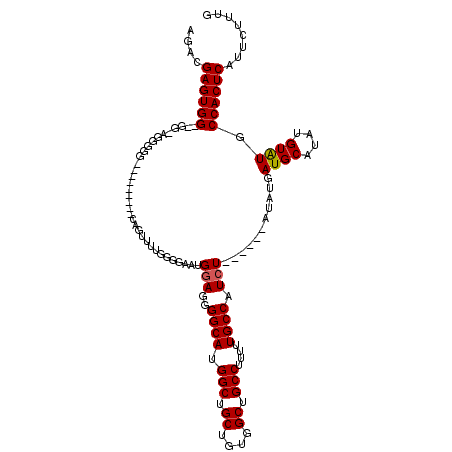
| Location | 22,305,199 – 22,305,313 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.24 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -25.26 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22305199 114 + 23771897 AGACGAGUGGCGGGGAGGGGGGUAAGGCUUCAGUUUUGGGGAAUGUAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU------AUAUGAUGCAUAUGUAUGCCACUCAUUCUUUG ....((((((((((((..((((.....))))..))))(.(..(((((((((((.(((.((....)).)))....)))).)))------))))..).).......))))))))........ ( -39.20) >DroSec_CAF1 63801 102 + 1 AGGCGAGUGG--GGCAGGGGG---------CAGUU-UGGCCAAUGGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU------AUAUGAUGCAUAUGUAUGCCACUCAUUCUUUG ....((((((--(((((((((---------(((((-.(((((.((......))))))).....))))))).)))))))....------.....((((....)))).))))))........ ( -38.30) >DroSim_CAF1 45840 103 + 1 AGGCGAGUGG--GGCAGGGGG---------CAGUUUUGGGCAAUGGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU------AUAUGAUGCAUAUGUAUGCCACUCAUUCUUUG ....((((((--(((((((((---------(((((..((((.(((......))).))).)...))))))).)))))))....------.....((((....)))).))))))........ ( -36.40) >DroEre_CAF1 62227 102 + 1 AGACGAGUGG--GG----------------CAGUUUUGGGGCUCGGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCUACUCGUAUAUGAUGCAUAUGUAUGCCACUCAUUCUUUG ..((((((((--((----------------(((((.....((((....))))..)))))))(((((.........)))))))))))))..((((.((((....))))...))))...... ( -34.60) >DroYak_CAF1 39464 95 + 1 AGACGAGUGG--GG----------------CAGUUUUGCGGAU-GGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU------AUAUGAUGCAUAUGUGUGCCACUCAUUCUUUG ....((((((--.(----------------((....((((.((-(((..((((.(((.((....)).)))....)))).)))------))....))))....))).))))))........ ( -38.60) >consensus AGACGAGUGG__GG_AGGGGG_________CAGUUUUGGGGAAUGGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU______AUAUGAUGCAUAUGUAUGCCACUCAUUCUUUG ....((((((..................................(((..((((.(((.((....)).)))....)))).)))...........((((....)))).))))))........ (-25.26 = -25.30 + 0.04)

| Location | 22,305,199 – 22,305,313 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.24 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22305199 114 - 23771897 CAAAGAAUGAGUGGCAUACAUAUGCAUCAUAU------AGAUGGCAAAAAGGCAGCCACAGCAGCCAUGCCCCUACAUUCCCCAAAACUGAAGCCUUACCCCCCUCCCCGCCACUCGUCU ...(((.((((((((.......((((((....------.))).)))..(((((.......(((....)))............((....))..)))))............))))))))))) ( -27.90) >DroSec_CAF1 63801 102 - 1 CAAAGAAUGAGUGGCAUACAUAUGCAUCAUAU------AGAUGGCAAAAAGGCAGCCACAGCAGCCAUGCCCCUCCAUUGGCCA-AACUG---------CCCCCUGCC--CCACUCGCCU ........((((((........((((((....------.))).)))....(((((.....((((....(((........)))..-..)))---------)...)))))--)))))).... ( -29.50) >DroSim_CAF1 45840 103 - 1 CAAAGAAUGAGUGGCAUACAUAUGCAUCAUAU------AGAUGGCAAAAAGGCAGCCACAGCAGCCAUGCCCCUCCAUUGCCCAAAACUG---------CCCCCUGCC--CCACUCGCCU ........(((((((((....)))).......------....((((....(((((........((.(((......))).))......)))---------))...))))--.))))).... ( -24.74) >DroEre_CAF1 62227 102 - 1 CAAAGAAUGAGUGGCAUACAUAUGCAUCAUAUACGAGUAGAUGGCAAAAAGGCAGCCACAGCAGCCAUGCCCCUCCGAGCCCCAAAACUG----------------CC--CCACUCGUCU ......((((...((((....)))).))))..((((((....((((....(((.((....)).)))..((........))........))----------------))--..)))))).. ( -24.30) >DroYak_CAF1 39464 95 - 1 CAAAGAAUGAGUGGCACACAUAUGCAUCAUAU------AGAUGGCAAAAAGGCAGCCACAGCAGCCAUGCCCCUCC-AUCCGCAAAACUG----------------CC--CCACUCGUCU ...(((.(((((((..((....((((((....------.)))((((....(((.((....)).))).)))).....-....)))....))----------------..--)))))))))) ( -25.60) >consensus CAAAGAAUGAGUGGCAUACAUAUGCAUCAUAU______AGAUGGCAAAAAGGCAGCCACAGCAGCCAUGCCCCUCCAUUCCCCAAAACUG_________CCCCCU_CC__CCACUCGUCU .......((((((((((....)))).................((((....(((.((....)).))).))))........................................))))))... (-20.24 = -20.44 + 0.20)


| Location | 22,305,239 – 22,305,353 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.07 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -30.86 |
| Energy contribution | -30.86 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22305239 114 + 23771897 GAAUGUAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU------AUAUGAUGCAUAUGUAUGCCACUCAUUCUUUGGCUCAUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUA ...(((.((((((.(((.((....)).))).....((((...------..(((((((....))).((((.........)))))))).......))))))))))..)))............ ( -29.30) >DroSec_CAF1 63829 114 + 1 CAAUGGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU------AUAUGAUGCAUAUGUAUGCCACUCAUUCUUUGGCCCCUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUA ....((((((((..(..((..((((((((.....(((.(((.------....))))))...))).))))).))..)....))))))))..(((((((......)).)))))......... ( -35.50) >DroSim_CAF1 45869 114 + 1 CAAUGGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU------AUAUGAUGCAUAUGUAUGCCACUCAUUCUUUGGCCCCUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUA ....((((((((..(..((..((((((((.....(((.(((.------....))))))...))).))))).))..)....))))))))..(((((((......)).)))))......... ( -35.50) >DroEre_CAF1 62249 120 + 1 GCUCGGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCUACUCGUAUAUGAUGCAUAUGUAUGCCACUCAUUCUUUGGCUCCUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUA ....((((((((..(..((..((((((((.....(((.(((...........))))))...))).))))).))..)....))))))))..(((((((......)).)))))......... ( -31.80) >DroYak_CAF1 39486 113 + 1 GAU-GGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU------AUAUGAUGCAUAUGUGUGCCACUCAUUCUUUGGCUCCUUCUUUUUUGGCUUCUCCGCACGAAACUAAAACUA (.(-((((((((..(((.((....)).))).....((((...------..((((.((((....))))...))))....)))).............))))))))))............... ( -32.40) >consensus GAAUGGAGGGGCAUGGCUGCUGUGGCUGCCUUUUUGCCAUCU______AUAUGAUGCAUAUGUAUGCCACUCAUUCUUUGGCUCCUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUA ....((((((((..(..((..((((((((.....(((.(((...........))))))...))).))))).))..)....))))))))..(((((((......)).)))))......... (-30.86 = -30.86 + -0.00)

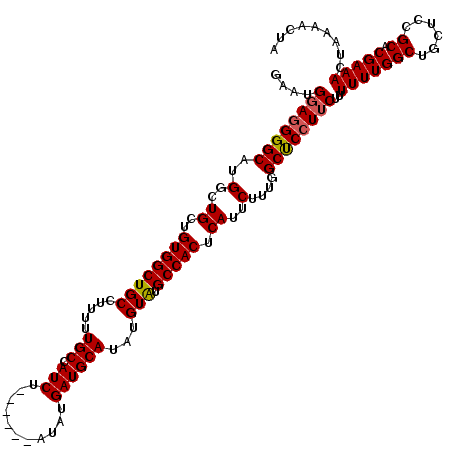

| Location | 22,305,313 – 22,305,433 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22305313 120 + 23771897 GCUCAUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUAAUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGCCAUCCAGCGUUUGCUCAUUUUUAGCGCAGGAACCU ((.((........)))).(((((((((((((...((((.....(((.............)))...))))..))))))))).))))..(((.(((((...........))))).))).... ( -29.92) >DroSec_CAF1 63903 120 + 1 GCCCCUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUAAUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGCCAUCCAGCGUUUGCUCAUUUUUAGCGCAGGAACCU (((...........))).(((((((((((((...((((.....(((.............)))...))))..))))))))).))))..(((.(((((...........))))).))).... ( -30.32) >DroSim_CAF1 45943 120 + 1 GCCCCUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUAAUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGCCAUCCAGCGUUUGCUCAUUUUUAGCGCAGGAACCU (((...........))).(((((((((((((...((((.....(((.............)))...))))..))))))))).))))..(((.(((((...........))))).))).... ( -30.32) >DroEre_CAF1 62329 116 + 1 GCUCCUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUAAUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGC----CAGCGUUUGCUCAUUUUUAGCGCAGGAACCU ..((((.(.......((((((((((((((((...((((.....(((.............)))...))))..))))))))).))).----))))....(((.......)))).)))).... ( -29.92) >DroYak_CAF1 39559 116 + 1 GCUCCUUCUUUUUUGGCUUCUCCGCACGAAACUAAAACUAAUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGC----CAGCGUUUGCUCAUUUUUAGCGCAGGAACCU ..((((.......((((((...(((((((((...((((.....(((.............)))...))))..))))))))).))))----))(((((...........))))))))).... ( -29.52) >consensus GCUCCUUCUUUUUUGGCUGCUCCGCACGAAACUAAAACUAAUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGCCAUCCAGCGUUUGCUCAUUUUUAGCGCAGGAACCU .....(((((...(((..(((((((((((((...((((.....(((.............)))...))))..))))))))).))))...)))(((((...........))))))))))... (-26.02 = -26.62 + 0.60)



| Location | 22,305,353 – 22,305,473 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.66 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -29.08 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22305353 120 + 23771897 AUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGCCAUCCAGCGUUUGCUCAUUUUUAGCGCAGGAACCUUUUUGUGCCAUUUUAUUUACAGGCAUGUAAACACAGGAGA ...........(((.((.....(((((((((...(((.(((((((.(........).)))).......((((((((....))))))))..........))).))).)))))))))))))) ( -30.20) >DroSec_CAF1 63943 120 + 1 AUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGCCAUCCAGCGUUUGCUCAUUUUUAGCGCAGGAACCUUUUUGUGCCAUUUUAUUUACAGGCAUGUAAACACACGAGA ...(((.............)))(((((((((...(((.(((((((.(........).)))).......((((((((....))))))))..........))).))).)))))))))..... ( -29.32) >DroSim_CAF1 45983 120 + 1 AUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGCCAUCCAGCGUUUGCUCAUUUUUAGCGCAGGAACCUUUUUGUGCCAUUUUAUUUACAGGCAUGUAAACACACGAGA ...(((.............)))(((((((((...(((.(((((((.(........).)))).......((((((((....))))))))..........))).))).)))))))))..... ( -29.32) >DroEre_CAF1 62369 116 + 1 AUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGC----CAGCGUUUGCUCAUUUUUAGCGCAGGAACCUUUUUGUGCCAUUUUAUUUACAGGCAUGUAAACACACGAGA ...(((.............)))(((((((((...(((.(((((((----........)))).......((((((((....))))))))..........))).))).)))))))))..... ( -30.12) >DroYak_CAF1 39599 116 + 1 AUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGC----CAGCGUUUGCUCAUUUUUAGCGCAGGAACCUUUUUGUGCCAUUUUAUUUACAGGCAUGUAAACACACGAGA ...(((.............)))(((((((((...(((.(((((((----........)))).......((((((((....))))))))..........))).))).)))))))))..... ( -30.12) >consensus AUUUGCAAAAAUCUACCACGCAUGUGUUUGCUUUUGUGUGUGAGCCAUCCAGCGUUUGCUCAUUUUUAGCGCAGGAACCUUUUUGUGCCAUUUUAUUUACAGGCAUGUAAACACACGAGA ...(((.............)))(((((((((...(((.((((((......(((....)))........((((((((....)))))))).......)))))).))).)))))))))..... (-29.08 = -29.08 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:41 2006