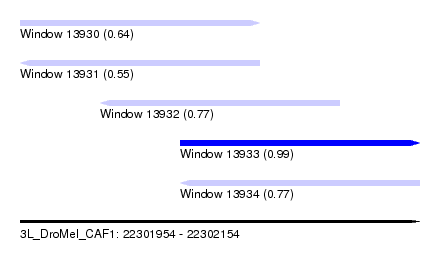
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,301,954 – 22,302,154 |
| Length | 200 |
| Max. P | 0.994988 |

| Location | 22,301,954 – 22,302,074 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -23.97 |
| Energy contribution | -24.01 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22301954 120 + 23771897 UCAUAUUAGUGGGAAAUGUAAUUUAACAUUGACAGAAGAACUUAUUAAUGUCAGCUCCCCCCUCUGUUUUACAAUAAAAGCGCACACAGAGUGCGCCAUUUUUUACGACACUUGGCUGCU ......(((.(((.(((((......)))))((((..............)))).......))).)))............(((((((.....))))((((..............)))).))) ( -24.18) >DroSec_CAF1 60581 118 + 1 UCAUAUUAGUGGGAAAUGUCAUUUAGCAUUGAGAGAAGAACUUAUUAAUGCCAGCUCCCGCC--UGUUUUACAAUAAAAGCGCACACUGAGUGCGCCAUUUUUUAUGACACUUGGCUGCU (((((...((((((..((.......((((((((((.....))).)))))))))..)))))).--...............((((((.....)))))).......)))))............ ( -28.51) >DroSim_CAF1 42646 118 + 1 UCAUAUUAGUGGGAAAUGUCAUUUAGCAUUGAGAGAAGAACUUAUUAAUGCCAGCUCCCGCC--UGUUUUACAAUAAAAGCGCACACUGAGUGCGCCAUUUUUUACGACACUUGGCUGCU ........((((((..((.......((((((((((.....))).)))))))))..)))))).--..............(((((((.....))))((((..............)))).))) ( -28.05) >DroEre_CAF1 59015 119 + 1 UCAUAUUGGGGGGAGAUGUCAUUUAGCACUGACAGAAGAACUUAUUAAUGUCAGCUCCC-CCUAUAUUUUACAAUAAAAGCGCACACUGAGUGCGCCAUUUUUUACGACACUUGGCUGCU ......(((((((((.(((......)))((((((..............)))))))))))-))))..............(((((((.....))))((((..............)))).))) ( -36.08) >DroYak_CAF1 36258 120 + 1 UCAUAUUAGGGGGAAAUGUCAUUUAGCAUUGACAGAAGAACUUAUUAAUGUCAGCUCCCCCCUCUGUUUAACAAUAAAAGCGCACGCUGAGUGCGCCAUUUUUUACGACACUUGGCUGCU ........(((((((((((......)))))((((..............))))...))))))....((((........))))(((.((..((((((..........)).))))..))))). ( -32.74) >consensus UCAUAUUAGUGGGAAAUGUCAUUUAGCAUUGACAGAAGAACUUAUUAAUGUCAGCUCCCCCCU_UGUUUUACAAUAAAAGCGCACACUGAGUGCGCCAUUUUUUACGACACUUGGCUGCU ........((((((..((.......(((((((.((......)).)))))))))..)))))).................(((((((.....))))((((..............)))).))) (-23.97 = -24.01 + 0.04)

| Location | 22,301,954 – 22,302,074 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22301954 120 - 23771897 AGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCUGUGUGCGCUUUUAUUGUAAAACAGAGGGGGGAGCUGACAUUAAUAAGUUCUUCUGUCAAUGUUAAAUUACAUUUCCCACUAAUAUGA ..((....((((...........(((((((.....))))))).....(((((((((((.(((((((((..........))))))))).))..))))...))))).....))))....)). ( -31.30) >DroSec_CAF1 60581 118 - 1 AGCAGCCAAGUGUCAUAAAAAAUGGCGCACUCAGUGUGCGCUUUUAUUGUAAAACA--GGCGGGAGCUGGCAUUAAUAAGUUCUUCUCUCAAUGCUAAAUGACAUUUCCCACUAAUAUGA ....(..(((((((((.......(((((((.....)))))))......(((....(--((..((((((..........))))))..)))...)))...)))))))))..).......... ( -28.60) >DroSim_CAF1 42646 118 - 1 AGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGUGUGCGCUUUUAUUGUAAAACA--GGCGGGAGCUGGCAUUAAUAAGUUCUUCUCUCAAUGCUAAAUGACAUUUCCCACUAAUAUGA (((((((...(((..((......(((((((.....))))))).......))..)))--)))(((((..(((........)))...)))))..))))........................ ( -28.32) >DroEre_CAF1 59015 119 - 1 AGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGUGUGCGCUUUUAUUGUAAAAUAUAGG-GGGAGCUGACAUUAAUAAGUUCUUCUGUCAGUGCUAAAUGACAUCUCCCCCCAAUAUGA ............(((((......(((((((.....)))))))................((-(((((.((.((((....(((.((......)).))).)))).)).)))))))...))))) ( -32.60) >DroYak_CAF1 36258 120 - 1 AGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGCGUGCGCUUUUAUUGUUAAACAGAGGGGGGAGCUGACAUUAAUAAGUUCUUCUGUCAAUGCUAAAUGACAUUUCCCCCUAAUAUGA .(((((..((((.(((........)))))))..)).)))...............((.(((((((((((..........)))))...(((((........)))))...))))))....)). ( -34.20) >consensus AGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGUGUGCGCUUUUAUUGUAAAACA_AGGCGGGAGCUGACAUUAAUAAGUUCUUCUGUCAAUGCUAAAUGACAUUUCCCACUAAUAUGA .......(((((((((.......(((((((.....)))))))..(((((.........((.(((((((..........)))))))))..)))))....)))))))))............. (-24.38 = -24.46 + 0.08)

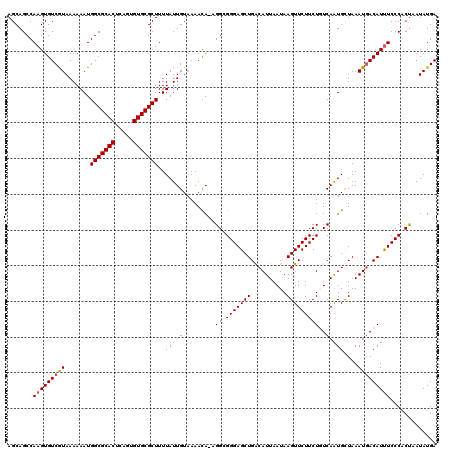
| Location | 22,301,994 – 22,302,114 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.66 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -26.38 |
| Energy contribution | -26.18 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22301994 120 - 23771897 CACUUUGAAGCAGCAGAGACAUAAAAAUGAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCUGUGUGCGCUUUUAUUGUAAAACAGAGGGGGGAGCUGACAUUAAUAAG ..(((((......)))))............(((((((.(.(((..((..(((((((.....))))))).((((((.((((.......))))..))))))..))..))).).))))))).. ( -30.60) >DroSec_CAF1 60621 118 - 1 CACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCAUAAAAAAUGGCGCACUCAGUGUGCGCUUUUAUUGUAAAACA--GGCGGGAGCUGGCAUUAAUAAG ..(((((......)))))............(((((((.(.(((.(((..((...((((.(((.(((((((.....)))))))))).))))...)).--)))....))).).))))))).. ( -29.20) >DroSim_CAF1 42686 118 - 1 CACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGUGUGCGCUUUUAUUGUAAAACA--GGCGGGAGCUGGCAUUAAUAAG ..(((((......)))))............(((((((.(.(((.(((...(((..((......(((((((.....))))))).......))..)))--)))....))).).))))))).. ( -29.42) >DroEre_CAF1 59055 119 - 1 CACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGUGUGCGCUUUUAUUGUAAAAUAUAGG-GGGAGCUGACAUUAAUAAG ..(((((......)))))............(((((((.(.(((..((..((((..((......(((((((.....))))))).......))..))))...-))..))).).))))))).. ( -28.52) >DroYak_CAF1 36298 120 - 1 CACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGCGUGCGCUUUUAUUGUUAAACAGAGGGGGGAGCUGACAUUAAUAAG ..(((((......)))))............(((((((.(.(((..((...(.((.(((.....(((((((.....))))))).......)))....)).).))..))).).))))))).. ( -26.30) >consensus CACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGUGUGCGCUUUUAUUGUAAAACA_AGGCGGGAGCUGACAUUAAUAAG ..(((((......)))))............(((((((.(.(((..((...(((..((......(((((((.....))))))).......))..))).....))..))).).))))))).. (-26.38 = -26.18 + -0.20)

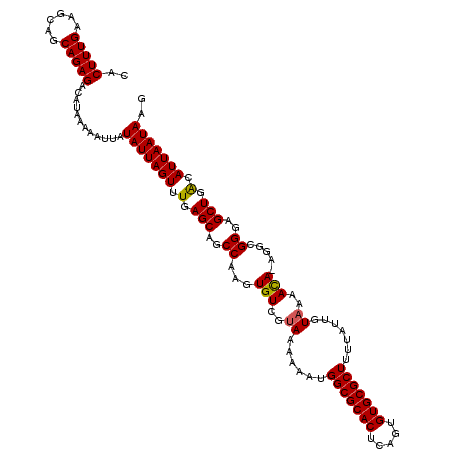
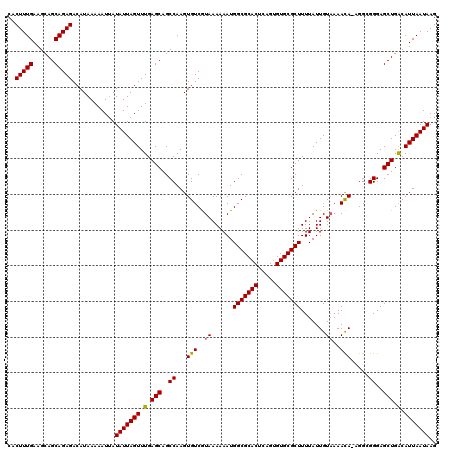
| Location | 22,302,034 – 22,302,154 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -32.48 |
| Energy contribution | -32.36 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22302034 120 + 23771897 CGCACACAGAGUGCGCCAUUUUUUACGACACUUGGCUGCUCAAACUAAUAUCAUUUUUAUGUCUCUGCUGCUUCAAAGUGUCAAUAAAAGUGCUGCGCCAAAAGUGGGACACAUUCCGGC .((.(((...(((((.(((((((...((((((((((.((....(((((........))).))....)).)))...)))))))...))))))).))))).....)))(((.....))).)) ( -30.10) >DroSec_CAF1 60659 120 + 1 CGCACACUGAGUGCGCCAUUUUUUAUGACACUUGGCUGCUCAAACUAAUAUAAUUUUUAUGUCUCUGCUGCUUCAAAGUGUCAAUAAAAGUGCUGCGCCAAAAGUGGGACACAUUCCGGC .((.((((..(((((.(((((((..(((((((((((.((....(((((........))).))....)).)))...))))))))..))))))).)))))....))))(((.....))).)) ( -34.10) >DroSim_CAF1 42724 120 + 1 CGCACACUGAGUGCGCCAUUUUUUACGACACUUGGCUGCUCAAACUAAUAUAAUUUUUAUGUCUCUGCUGCUUCAAAGUGUCAAUAAAAGUGCUGCGCCAAAAGUGGGACACAUUCCGGC .((.((((..(((((.(((((((...((((((((((.((....(((((........))).))....)).)))...)))))))...))))))).)))))....))))(((.....))).)) ( -32.90) >DroEre_CAF1 59094 120 + 1 CGCACACUGAGUGCGCCAUUUUUUACGACACUUGGCUGCUCAAACUAAUAUAAUUUUUAUGUCUCUGCUGCUUCAAAGUGUCAAUAAAAGUGCUGCGCCAAAAGUGGGACACAUUCCGGC .((.((((..(((((.(((((((...((((((((((.((....(((((........))).))....)).)))...)))))))...))))))).)))))....))))(((.....))).)) ( -32.90) >DroYak_CAF1 36338 120 + 1 CGCACGCUGAGUGCGCCAUUUUUUACGACACUUGGCUGCUCAAACUAAUAUAAUUUUUAUGUCUCUGCUGCUUCAAAGUGUCAAUAAAAAUGCUGCGCCAAAAGUGGGACACAUUCCGGC .((.((((..(((((.(((((((...((((((((((.((....(((((........))).))....)).)))...)))))))...))))))).)))))....))))(((.....))).)) ( -32.50) >consensus CGCACACUGAGUGCGCCAUUUUUUACGACACUUGGCUGCUCAAACUAAUAUAAUUUUUAUGUCUCUGCUGCUUCAAAGUGUCAAUAAAAGUGCUGCGCCAAAAGUGGGACACAUUCCGGC .((.((((..(((((.(((((((...((((((((((.((....(((((........))).))....)).)))...)))))))...))))))).)))))....))))(((.....))).)) (-32.48 = -32.36 + -0.12)

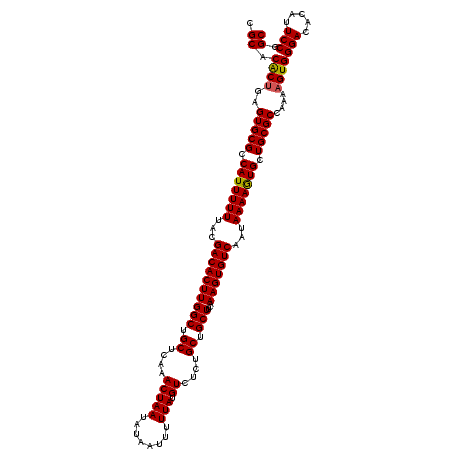
| Location | 22,302,034 – 22,302,154 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -33.02 |
| Energy contribution | -32.86 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22302034 120 - 23771897 GCCGGAAUGUGUCCCACUUUUGGCGCAGCACUUUUAUUGACACUUUGAAGCAGCAGAGACAUAAAAAUGAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCUGUGUGCG (((((((.(((...))))))))))((.....((((((.(((((((....((.((..((((.(((........)))))))..)).)).)))))))))))))....))((((.....)))). ( -32.90) >DroSec_CAF1 60659 120 - 1 GCCGGAAUGUGUCCCACUUUUGGCGCAGCACUUUUAUUGACACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCAUAAAAAAUGGCGCACUCAGUGUGCG (((((((.(((...))))))))))((.....((((((.(((((((....((.((..((((.(((........)))))))..)).)).)))))))))))))....))((((.....)))). ( -33.30) >DroSim_CAF1 42724 120 - 1 GCCGGAAUGUGUCCCACUUUUGGCGCAGCACUUUUAUUGACACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGUGUGCG (((((((.(((...))))))))))((.....((((((.(((((((....((.((..((((.(((........)))))))..)).)).)))))))))))))....))((((.....)))). ( -32.90) >DroEre_CAF1 59094 120 - 1 GCCGGAAUGUGUCCCACUUUUGGCGCAGCACUUUUAUUGACACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGUGUGCG (((((((.(((...))))))))))((.....((((((.(((((((....((.((..((((.(((........)))))))..)).)).)))))))))))))....))((((.....)))). ( -32.90) >DroYak_CAF1 36338 120 - 1 GCCGGAAUGUGUCCCACUUUUGGCGCAGCAUUUUUAUUGACACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGCGUGCG (((((((.(((...))))))))))((..(((((((..((((((((....((.((..((((.(((........)))))))..)).)).))))))))..)))))))((((.....)))))). ( -35.20) >consensus GCCGGAAUGUGUCCCACUUUUGGCGCAGCACUUUUAUUGACACUUUGAAGCAGCAGAGACAUAAAAAUUAUAUUAGUUUGAGCAGCCAAGUGUCGUAAAAAAUGGCGCACUCAGUGUGCG (((((((.(((...))))))))))((.....((((((.(((((((....((.((..((((.(((........)))))))..)).)).)))))))))))))....))((((.....)))). (-33.02 = -32.86 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:33 2006