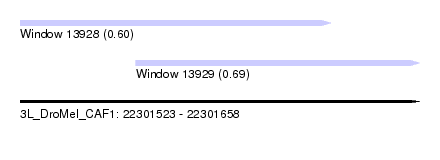
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,301,523 – 22,301,658 |
| Length | 135 |
| Max. P | 0.691655 |

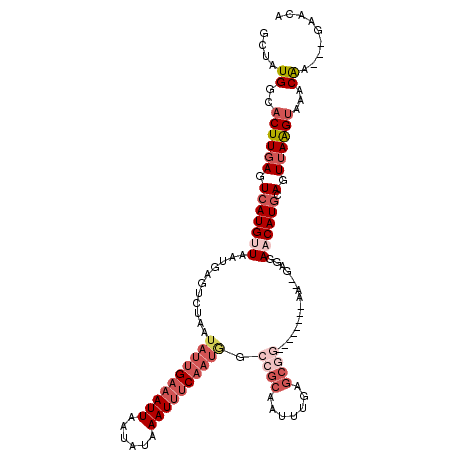
| Location | 22,301,523 – 22,301,628 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -17.62 |
| Energy contribution | -19.82 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22301523 105 + 23771897 ACUUCGUACACCCUGCAGCCUGGCAUCCC-GGGGCUUUUAGCUAUGGCACUUGAGUCAUGUUAAUGAGUCUAAUAUUGAAAUUAAUAUAAAUAUCAAUAGCCGCAA-------------- .............(((..(((((....))-)))((.....))...(((....((.((((....)))).))...((((((.(((......))).)))))))))))).-------------- ( -25.20) >DroSec_CAF1 60128 119 + 1 ACUUCGUACACCCUGCAGCCUGGCAUCCC-GUGGCUUUUAGCUAUGUCACUUGAGUCAUGUUAAUGAGUCUAAUAUUGAAAUUAAUAUAAAUUUCAAUGGCCGCAAUUUGAGCGGUGAGG ........((((((.(((..((((....(-(((((.....))))))......((.((((....)))).))...((((((((((......))))))))))))))....))))).))))... ( -31.20) >DroSim_CAF1 42193 119 + 1 ACUUCGUACACCCUGCAGCCUGGCAUCCC-GUGGCUUUUAGCUAUGGCACUUGAGUCAUGUUAAUGAGUCUAAUAUUGAAAUUAAUAUAAAUUUCAAUGGCCGCAAUUUGAGCGGUGAGG ........((((((.(((..((((...((-(((((.....))))))).....((.((((....)))).))...((((((((((......))))))))))))))....))))).))))... ( -34.30) >DroEre_CAF1 58633 95 + 1 -CUUCCUACACCCUGCCGCCUGGUAGCCC-GGGGCUUUUAGCUAUGGCACUUGAGUCAUGCUAAUGAGUCUAGUAUUGCAAUUAAUAUAAAUUUC------------------GG----- -...........(((((....))))).((-((((((((((((.(((((......)))))))))).)))))).((((((....))))))......)------------------))----- ( -24.90) >DroYak_CAF1 24920 114 + 1 -CUUCCUACACCCUGCCGCCUGGCCUCCCAGGGACUUUUAGCUAUGGCACUUUAGUCAUGCUAAUUAGUCCAAUAUUGAAAUUAAUACAAAUUUCAAUGCCCGCAAUUUGAGCGG----- -..............((((((((....))))(((((.(((((.(((((......))))))))))..)))))..((((((((((......))))))))))............))))----- ( -34.50) >consensus ACUUCGUACACCCUGCAGCCUGGCAUCCC_GGGGCUUUUAGCUAUGGCACUUGAGUCAUGUUAAUGAGUCUAAUAUUGAAAUUAAUAUAAAUUUCAAUGGCCGCAAUUUGAGCGG_____ .....................(((.......(((((((((((.(((((......)))))))))).))))))..((((((((((......))))))))))))).................. (-17.62 = -19.82 + 2.20)

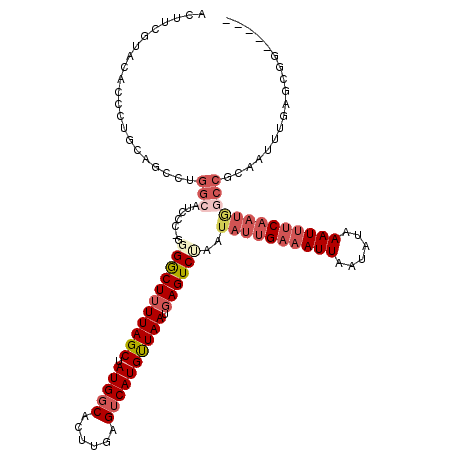
| Location | 22,301,562 – 22,301,658 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.12 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -10.50 |
| Energy contribution | -13.56 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22301562 96 + 23771897 GCUAUGGCACUUGAGUCAUGUUAAUGAGUCUAAUAUUGAAAUUAAUAUAAAUAUCAAUAGCCGCAA---------------------GAGAAACAUGCAGUUAAGUAAACAA---GAACA (((..(((....((.((((....)))).))...((((((.(((......))).)))))))))(((.---------------------(.....).))).....)))......---..... ( -17.50) >DroSec_CAF1 60167 117 + 1 GCUAUGUCACUUGAGUCAUGUUAAUGAGUCUAAUAUUGAAAUUAAUAUAAAUUUCAAUGGCCGCAAUUUGAGCGGUGAGGUGGCAGAGGAGAACAUACAGUUAAGUAAACGA---GAACA .((.((((((((((.((((....)))).))...((((((((((......))))))))))(((((.......))))).)))))))).))...........(((..(....)..---.))). ( -31.30) >DroSim_CAF1 42232 117 + 1 GCUAUGGCACUUGAGUCAUGUUAAUGAGUCUAAUAUUGAAAUUAAUAUAAAUUUCAAUGGCCGCAAUUUGAGCGGUGAGGGGGCAGAGGAGAACAUGCAGUUAAGUAAACAA---GAACA ((....))((((((.(((((((.....((((..((((((((((......))))))))))(((((.......)))))....)))).......)))))).).))))))......---..... ( -30.30) >DroEre_CAF1 58671 89 + 1 GCUAUGGCACUUGAGUCAUGCUAAUGAGUCUAGUAUUGCAAUUAAUAUAAAUUUC------------------GG--------AA--GAGGAACAUGCAGUUAAGUACACAA---GAACA ((.((((((((.((.((((....)))).)).)))...))............((((------------------..--------..--))))..)))))..............---..... ( -16.30) >DroYak_CAF1 24959 110 + 1 GCUAUGGCACUUUAGUCAUGCUAAUUAGUCCAAUAUUGAAAUUAAUACAAAUUUCAAUGCCCGCAAUUUGAGCGG--------AG--GAGGAACAUGCAGUUAGGCAAACAAGAAGAACA ((((((((......)))))((...((..(((..((((((((((......)))))))))).((((.......))))--------.)--))..))...)).....))).............. ( -25.60) >consensus GCUAUGGCACUUGAGUCAUGUUAAUGAGUCUAAUAUUGAAAUUAAUAUAAAUUUCAAUGGCCGCAAUUUGAGCGG________AA__GAGGAACAUGCAGUUAAGUAAACAA___GAACA ....((..((((((.(((((((...........((((((((((......)))))))))).((((.......))))................)))))).).))))))...))......... (-10.50 = -13.56 + 3.06)

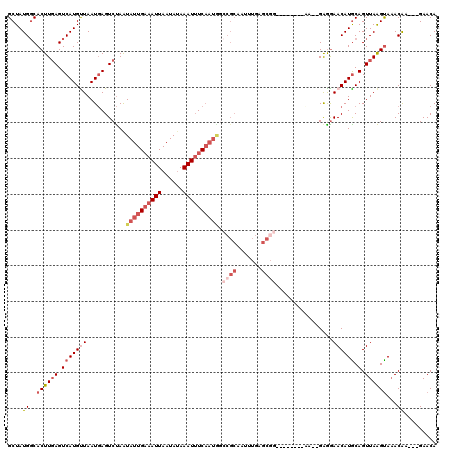
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:28 2006