
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
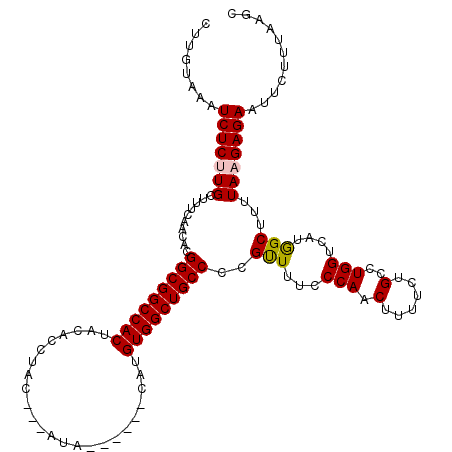
| Location | 22,299,161 – 22,299,272 |
| Length | 111 |
| Max. P | 0.999705 |

| Location | 22,299,161 – 22,299,272 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.86 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.92 |
| SVM RNA-class probability | 0.999705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22299161 111 + 23771897 CUUGUAAAUCUCUUGCUUUCAACACGGCGGCCACUACACCUAC---AUA------CAUGUGGCUGCCCCGUUUCCCCAACUUUUCUGCCUGGUCAUGGCUUUUAAGAGAAUUCUUUAAGC ........(((((((.....(((..(((((((((.........---...------...)))))))))..)))..............(((.......)))...)))))))........... ( -28.56) >DroSec_CAF1 57720 111 + 1 CUUGUAAAUCUCCUGCUUUCAACACGGCGGCCACUACACCUAC---AUA------CAUGUGGCUGCCCCGCUUUACCAACUUUUCUGCCUGGUCAUGGCUUUUAAGAGAAUUCUUUAAGC ........((((.............(((((((((.........---...------...)))))))))..(((..((((.(......)..))))...)))......))))........... ( -26.96) >DroSim_CAF1 39794 111 + 1 CUUGUAAAUCUCCUGCUUUCAACACGGCGGCCACUACACCUAC---AUA------CAUGUGGCUGCCCCGCUUUACCAACUUUUCUGCCUGGUCAUGGCUUUUAAGAGAAUUCUUUAAGC ........((((.............(((((((((.........---...------...)))))))))..(((..((((.(......)..))))...)))......))))........... ( -26.96) >DroEre_CAF1 56251 120 + 1 CUUGUAAAUCUCUUGCUUUCAACACGGCGGCCACCACACCUACUACCUACUGCCCCGUGUGGCUGCCCCGUUUUCCCAACUUUUCUGCCUGGUCAUAGCUUUUAAGAGAAUUCUUUAAGC ........(((((((((.....((.((((((..((((((.................))))))..)))..(((.....)))......))))).....)))....))))))........... ( -26.13) >DroYak_CAF1 22534 107 + 1 CUUGUAAAUCUCUUGCUUUCAACACGGCGGCCACUACACCUAC-------------AUGUGGCUGCCCCGUUUUCCCAACUUUUCUGCCUGGUCAUAGCUUUUAAGAGAAUUCUUUAAGC ........(((((((..........(((((((((.........-------------..)))))))))..(((...(((.(......)..)))....)))...)))))))........... ( -26.20) >consensus CUUGUAAAUCUCUUGCUUUCAACACGGCGGCCACUACACCUAC___AUA______CAUGUGGCUGCCCCGUUUUCCCAACUUUUCUGCCUGGUCAUGGCUUUUAAGAGAAUUCUUUAAGC ........(((((((..........(((((((((........................)))))))))..(((...(((.(......)..)))....)))...)))))))........... (-24.94 = -24.86 + -0.08)

| Location | 22,299,161 – 22,299,272 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -29.36 |
| Energy contribution | -28.64 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22299161 111 - 23771897 GCUUAAAGAAUUCUCUUAAAAGCCAUGACCAGGCAGAAAAGUUGGGGAAACGGGGCAGCCACAUG------UAU---GUAGGUGUAGUGGCCGCCGUGUUGAAAGCAAGAGAUUUACAAG ...........((((((....(((.......)))......(((.....((((.(((.((((((..------(..---....)..).))))).))).))))...)))))))))........ ( -31.20) >DroSec_CAF1 57720 111 - 1 GCUUAAAGAAUUCUCUUAAAAGCCAUGACCAGGCAGAAAAGUUGGUAAAGCGGGGCAGCCACAUG------UAU---GUAGGUGUAGUGGCCGCCGUGUUGAAAGCAGGAGAUUUACAAG ...........((((((....(((.......)))......(((.....((((.(((.((((((..------(..---....)..).))))).))).))))...)))))))))........ ( -30.90) >DroSim_CAF1 39794 111 - 1 GCUUAAAGAAUUCUCUUAAAAGCCAUGACCAGGCAGAAAAGUUGGUAAAGCGGGGCAGCCACAUG------UAU---GUAGGUGUAGUGGCCGCCGUGUUGAAAGCAGGAGAUUUACAAG ...........((((((....(((.......)))......(((.....((((.(((.((((((..------(..---....)..).))))).))).))))...)))))))))........ ( -30.90) >DroEre_CAF1 56251 120 - 1 GCUUAAAGAAUUCUCUUAAAAGCUAUGACCAGGCAGAAAAGUUGGGAAAACGGGGCAGCCACACGGGGCAGUAGGUAGUAGGUGUGGUGGCCGCCGUGUUGAAAGCAAGAGAUUUACAAG ...........((((((....(((....((..((......))..))...(((((((.(((((((...((........))..))))))).))).))))......)))))))))........ ( -35.40) >DroYak_CAF1 22534 107 - 1 GCUUAAAGAAUUCUCUUAAAAGCUAUGACCAGGCAGAAAAGUUGGGAAAACGGGGCAGCCACAU-------------GUAGGUGUAGUGGCCGCCGUGUUGAAAGCAAGAGAUUUACAAG ...........((((((....(((....((..((......))..))..((((.(((.(((((((-------------......)).))))).))).))))...)))))))))........ ( -29.30) >consensus GCUUAAAGAAUUCUCUUAAAAGCCAUGACCAGGCAGAAAAGUUGGGAAAACGGGGCAGCCACAUG______UAU___GUAGGUGUAGUGGCCGCCGUGUUGAAAGCAAGAGAUUUACAAG ...........((((((....(((.......)))......(((.....((((.(((.(((((........................))))).))).))))...)))))))))........ (-29.36 = -28.64 + -0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:25 2006