
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,295,128 – 22,295,239 |
| Length | 111 |
| Max. P | 0.811174 |

| Location | 22,295,128 – 22,295,239 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -29.59 |
| Energy contribution | -30.90 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
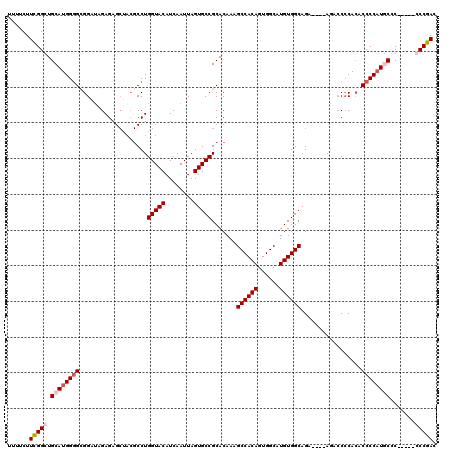
>3L_DroMel_CAF1 22295128 111 + 23771897 UCUUCUUCGGCUGCAUGGGGCGGAUAGAGAGCUACGCCUGGUACAUCAAUUAGUGCCGCACAAAGCCACAGUGGCAUGUGGCAGA----AGACCCCACACCCCAUGCAC-----CCCGAC ......((((.(((((((((.(.............((..(((((........))))))).....((((((......))))))...----........).))))))))).-----.)))). ( -41.50) >DroSec_CAF1 53665 110 + 1 UUUUCUUCGGCUGCAUGGGGCGGAUAGAGAGCUACGCCUGGUACAUCAAUUAGUGCCGCACAAAGCCACAGUGGCAUGUGGCAGA----AGACCACACACCCCUUGCUC------CCGAC ......((((..(((.((((.(........(((((....(((((........)))))((.....))....))))).((((((...----.).)))))).)))).)))..------)))). ( -40.30) >DroEre_CAF1 52376 120 + 1 UUUUCUUCGGGUGCAUGGGGUGGAGGGAGAGCAACGCCUGGUACAUCAAUUAGUGCCGCACAAAGCCACAGUGGCAUGUGGCAGACCCCACACCCCACACCCCAUCCCCCUUUGCCCGAC ......(((((((.((((((((..(((........((..(((((........))))))).....((((((......))))))..........)))..)))))))).)......)))))). ( -48.00) >DroYak_CAF1 18305 105 + 1 UUUUCUUUGGGUGCAUGG-GCGGAUAGAGAG-----CCUGGUACAUCAAUUAGUGCCGCACAAAGCCACAGUGGCAUGUGGCAGG----AGAACCCAAACCCCAUUCCC-----CCCGAC .....(((((((((..((-((.........)-----)))(((((........))))))).....((((((......))))))...----...)))))))..........-----...... ( -33.50) >consensus UUUUCUUCGGCUGCAUGGGGCGGAUAGAGAGCUACGCCUGGUACAUCAAUUAGUGCCGCACAAAGCCACAGUGGCAUGUGGCAGA____AGACCCCACACCCCAUGCCC_____CCCGAC ......(((((.((((((((...................(((((........))))).......((((((......)))))).................)))))))).......))))). (-29.59 = -30.90 + 1.31)


| Location | 22,295,128 – 22,295,239 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -44.18 |
| Consensus MFE | -30.70 |
| Energy contribution | -32.89 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
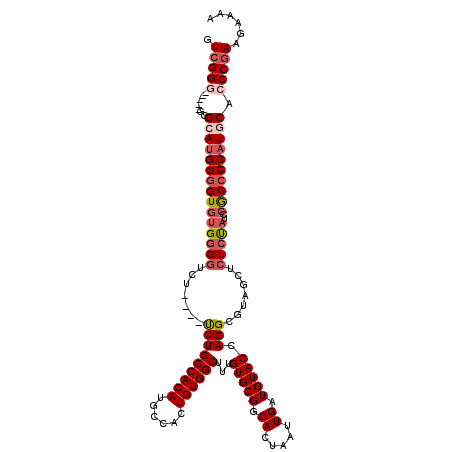
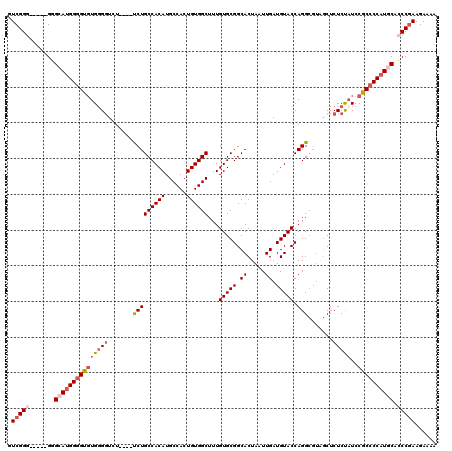
>3L_DroMel_CAF1 22295128 111 - 23771897 GUCGGG-----GUGCAUGGGGUGUGGGGUCU----UCUGCCACAUGCCACUGUGGCUUUGUGCGGCACUAAUUGAUGUACCAGGCGUAGCUCUCUAUCCGCCCCAUGCAGCCGAAGAAGA ....((-----.((((((((((((((((...----.((((((((......))))((((.(((((.((.....)).))))).))))))))..)))))..))))))))))).))........ ( -46.10) >DroSec_CAF1 53665 110 - 1 GUCGG------GAGCAAGGGGUGUGUGGUCU----UCUGCCACAUGCCACUGUGGCUUUGUGCGGCACUAAUUGAUGUACCAGGCGUAGCUCUCUAUCCGCCCCAUGCAGCCGAAGAAAA .((((------..(((.((((((..(((...----.((((((((......))))((((.(((((.((.....)).))))).))))))))....)))..)))))).)))..))))...... ( -42.20) >DroEre_CAF1 52376 120 - 1 GUCGGGCAAAGGGGGAUGGGGUGUGGGGUGUGGGGUCUGCCACAUGCCACUGUGGCUUUGUGCGGCACUAAUUGAUGUACCAGGCGUUGCUCUCCCUCCACCCCAUGCACCCGAAGAAAA ..........((((.((((((((..(((.(..(.((((((((((......))))))...(((((.((.....)).))))).)))).)..)...)))..)))))))).).)))........ ( -52.90) >DroYak_CAF1 18305 105 - 1 GUCGGG-----GGGAAUGGGGUUUGGGUUCU----CCUGCCACAUGCCACUGUGGCUUUGUGCGGCACUAAUUGAUGUACCAGG-----CUCUCUAUCCGC-CCAUGCACCCAAAGAAAA ...(((-----.(((..(((((((((..((.----..((((.((((((.....)))...))).))))......))....)))))-----))))...))).)-))................ ( -35.50) >consensus GUCGGG_____GGGCAUGGGGUGUGGGGUCU____UCUGCCACAUGCCACUGUGGCUUUGUGCGGCACUAAUUGAUGUACCAGGCGUAGCUCUCUAUCCGCCCCAUGCACCCGAAGAAAA .(((((.......(((((((((((((((.......(((((((((......))))))...(((((.((.....)).))))).))).......)))))..)))))))))).)))))...... (-30.70 = -32.89 + 2.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:17 2006