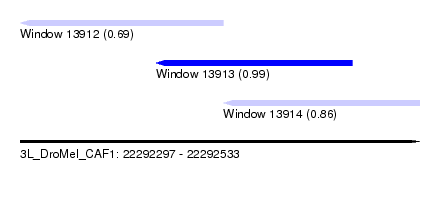
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,292,297 – 22,292,533 |
| Length | 236 |
| Max. P | 0.989789 |

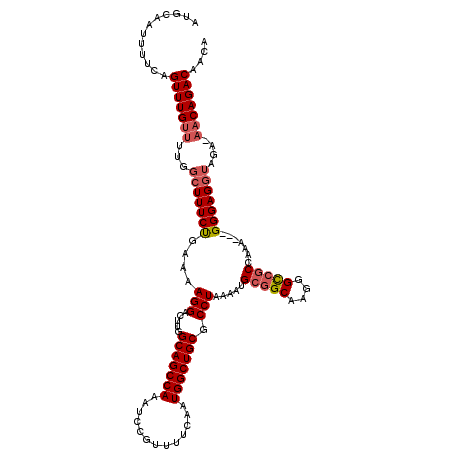
| Location | 22,292,297 – 22,292,417 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -23.24 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22292297 120 - 23771897 AAGUCGUAAACAUUAUUUCUAAAAAUUGCUUUUGUCGCAUUUCCUCGACAUAUUUGGCAGCAGUUCAAGACGCCAGAGCUGCUCUGGCGAACAAUUCCCACCACUUAGGAGGACAUAAAC ..((((....)....(((((((...(((((..(((((........))))).....)))))..........((((((((...))))))))...............))))))))))...... ( -28.50) >DroSec_CAF1 50767 120 - 1 AAGUCGUAAACAUUAUUCCUAAAAAUUGCUUUUGUCGCAUUUCCUCGACAUAUUUGGCAGGAGUUCAAGACGCCAGAGCUGCUCUGGCCAACAAUUCCCACCACUUAGGAGAACAUAAAC ...............(((((((....((((..(((((........))))).....))))((((((......(((((((...)))))))....))))))......)))))))......... ( -30.50) >DroEre_CAF1 49126 117 - 1 GAGUCGUAAACAUUAUUUCUAAAAAUAGCUUUUGUCGCAUUUUCUCGACACAAUUGGCAGCAGCUCAAGACGCCAGAG---CUCUGGCCAACAAUGCCCACCACUUAGAAGAACAUAAAC ...............(((((((..........(((((........)))))...(((((((.(((((.........)))---)))).))))).............)))))))......... ( -22.90) >DroYak_CAF1 15160 117 - 1 GAGUCGUAAACAUUAUUUCUAAAAAUUGCUUUUGUCGCAUUUUCUCGACACAUUUGGCAGCAGUUCAAGACGCCAGAA---CUCUGGCCAACAAUUCUUACCACUUAGAAGAACAUAAAC .........................(((((..(((((........))))).....)))))..((((.....(((((..---..)))))......((((........))))))))...... ( -22.50) >consensus AAGUCGUAAACAUUAUUUCUAAAAAUUGCUUUUGUCGCAUUUCCUCGACACAUUUGGCAGCAGUUCAAGACGCCAGAG___CUCUGGCCAACAAUUCCCACCACUUAGAAGAACAUAAAC ...............(((((((...(((((..(((((........))))).....)))))...........(((((((...)))))))................)))))))......... (-23.24 = -23.30 + 0.06)

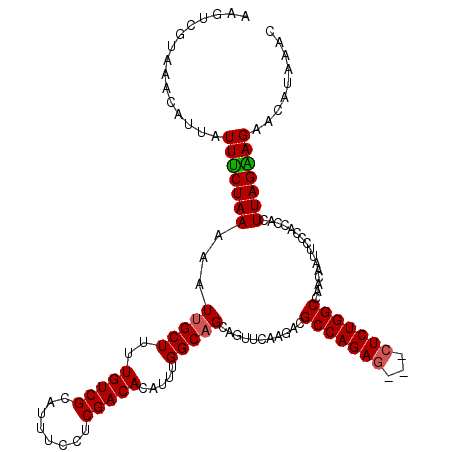

| Location | 22,292,377 – 22,292,493 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -33.78 |
| Energy contribution | -33.65 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22292377 116 - 23771897 UGCAGCCAAAUCCGUUUUCGAUGGCUGCGCCUAAAGUGCGGCAAGGGCAGCCAA----GGGAGGUAGACAACAGACAGCAAAGUCGUAAACAUUAUUUCUAAAAAUUGCUUUUGUCGCAU .(((((((....((....)).))))))).......(((((((((((((((....----.((((((((......(((......))).......)))))))).....))))))))))))))) ( -41.72) >DroSec_CAF1 50847 120 - 1 GGCAGCCAAAUCCGUUUUCGAUGGCUGCGCCUAAAGUGCGGCAAGGGUCACCAAAGGGGGGAGGUAGAAAACAGACAGCAAAGUCGUAAACAUUAUUCCUAAAAAUUGCUUUUGUCGCAU .(((((((....((....)).))))))).......(((((((((((((..((....))(((((..........(((......)))..........))))).......))))))))))))) ( -38.25) >DroEre_CAF1 49203 110 - 1 GGCAGCCAAAUCCGUUUUCAAUGGCUGCGCCUAAAAUGCGGCAAUGGCCGCCAGA---GGGAGC-------CAGACAACAGAGUCGUAAACAUUAUUUCUAAAAAUAGCUUUUGUCGCAU .(((((((.............))))))).......(((((((((((((..((...---.)).))-------))(((......)))..........................))))))))) ( -32.62) >DroYak_CAF1 15237 117 - 1 GGCAGCCAAAUCCGUUUUCAAUGGCUGCGCCUAAAAUGCGGCAAUGGCCGCCACC---GGGAGGUAGAUAACAGACAACAGAGUCGUAAACAUUAUUUCUAAAAAUUGCUUUUGUCGCAU .(((((((.............))))))).......(((((((((.(((.......---.((((((((......(((......))).......)))))))).......))).))))))))) ( -34.90) >consensus GGCAGCCAAAUCCGUUUUCAAUGGCUGCGCCUAAAAUGCGGCAAGGGCCGCCAAA___GGGAGGUAGA_AACAGACAACAAAGUCGUAAACAUUAUUUCUAAAAAUUGCUUUUGUCGCAU .(((((((.............))))))).......(((((((((((((..........(((((..........(((......)))..........))))).......))))))))))))) (-33.78 = -33.65 + -0.13)

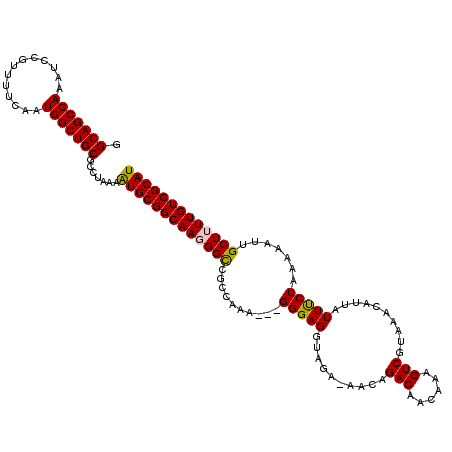
| Location | 22,292,417 – 22,292,533 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -29.90 |
| Energy contribution | -31.02 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22292417 116 - 23771897 AUGCAAUUUUCAGUUUGUUUUGGCUUUCUGAAAAGGACUUUGCAGCCAAAUCCGUUUUCGAUGGCUGCGCCUAAAGUGCGGCAAGGGCAGCCAA----GGGAGGUAGACAACAGACAGCA ............(((((((...(((((((((((((((.((((....))))))).)))))..((((((((((........)))....))))))).----)))))))....))))))).... ( -37.20) >DroSec_CAF1 50887 120 - 1 AUGCAAUUUUCAGUUUGUUUUGGCUUUCUGAAAAGGACUUGGCAGCCAAAUCCGUUUUCGAUGGCUGCGCCUAAAGUGCGGCAAGGGUCACCAAAGGGGGGAGGUAGAAAACAGACAGCA ............(((((((((.(((((((((((((((.((((...)))).))).)))))(((.(((((((.....)))))))....))).((....)))))))))..))))))))).... ( -39.50) >DroEre_CAF1 49243 110 - 1 AUGCAAUUUUCAGUUUGUUUUGGCUUUCCGAACAGGACUUGGCAGCCAAAUCCGUUUUCAAUGGCUGCGCCUAAAAUGCGGCAAUGGCCGCCAGA---GGGAGC-------CAGACAACA ............(((.((((.((((((((.....)))....(((((((.............))))))).(((.....(((((....)))))...)---))))))-------)))))))). ( -37.22) >DroYak_CAF1 15277 117 - 1 AUGCAAUUUUCAGUUUGUUUUGGCUUUCUGAACAGGACUUGGCAGCCAAAUCCGUUUUCAAUGGCUGCGCCUAAAAUGCGGCAAUGGCCGCCACC---GGGAGGUAGAUAACAGACAACA ............(((((((...((((((((...(((.....(((((((.............))))))).))).....(((((....)))))...)---)))))))....))))))).... ( -39.22) >consensus AUGCAAUUUUCAGUUUGUUUUGGCUUUCUGAAAAGGACUUGGCAGCCAAAUCCGUUUUCAAUGGCUGCGCCUAAAAUGCGGCAAGGGCCGCCAAA___GGGAGGUAGA_AACAGACAACA ............(((((((...(((((((....(((.....(((((((.............))))))).))).....(((((....))))).......)))))))....))))))).... (-29.90 = -31.02 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:13 2006