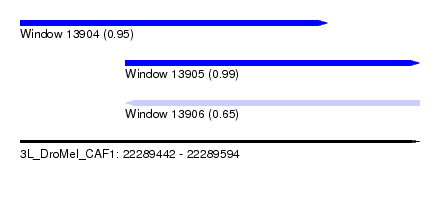
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,289,442 – 22,289,594 |
| Length | 152 |
| Max. P | 0.993421 |

| Location | 22,289,442 – 22,289,559 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -30.49 |
| Energy contribution | -31.49 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
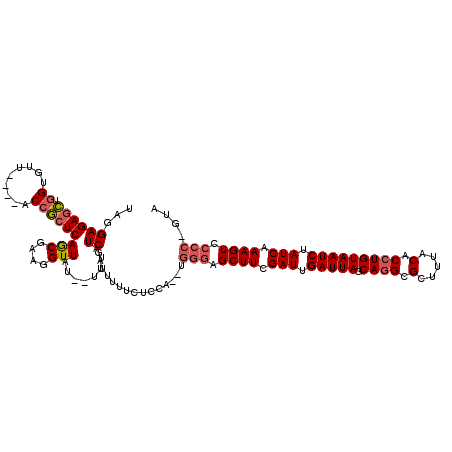
>3L_DroMel_CAF1 22289442 117 + 23771897 GUUGUUUGUUAUGGUAACGAGUGUAAAUUUCAUGGCAAAUUACCGGGAACUUUGAUAGAUUACAGGUGUAAAGCGCCUGCUAAUCAAUCGAAGCUCCCA--UGGAGAAAAAUCGAUAAA- ((.(((((((((((...............))))))))))).)).((((.(((((((.(((((((((((.....)))))).))))).))))))).))))(--(.((......)).))...- ( -40.26) >DroSec_CAF1 47846 117 + 1 GUUGUUUGUUAUGGUAACGAGUGUAAAUUUCAUGGCAAUUUAC-GGGGACUUUGAUAGAUUACAGGUGUAAAGCGCCUGCUAAUCAAUCGAAGCUCCCA--UGGUGAAAAAUCGAUAAAA ((...(((((((((...............)))))))))...))-(((..(((((((.(((((((((((.....)))))).))))).)))))))..))).--((((.....))))...... ( -38.76) >DroEre_CAF1 46337 118 + 1 GUUGUUUGUUAUGGUAACGAGCGUACAUUUCAUGGCAAUUUAC-GGGCACUUUGAUAGAUUACAGGUGUAAAACGCUUGCUAAUCAAUCGAAGCUGGGAGCUAGAGAAAAAUCGAUGAA- (((((((((((...))))))))).)).((((.((((......(-.(((..((((((.(((((((((((.....)))))).))))).))))))))).)..))))))))............- ( -31.60) >consensus GUUGUUUGUUAUGGUAACGAGUGUAAAUUUCAUGGCAAUUUAC_GGGAACUUUGAUAGAUUACAGGUGUAAAGCGCCUGCUAAUCAAUCGAAGCUCCCA__UGGAGAAAAAUCGAUAAA_ ((((.(((((((((...............)))))))))......((((.(((((((.(((((((((((.....)))))).))))).))))))).))))..............)))).... (-30.49 = -31.49 + 1.00)

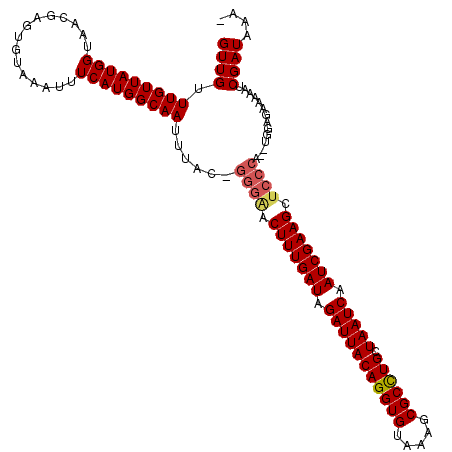

| Location | 22,289,482 – 22,289,594 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -27.53 |
| Energy contribution | -27.87 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.15 |
| Mean z-score | -4.38 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22289482 112 + 23771897 UACCGGGAACUUUGAUAGAUUACAGGUGUAAAGCGCCUGCUAAUCAAUCGAAGCUCCCA--UGGAGAAAAAUCGAUAAA--AUAACCUUCGCUGAGCGGU----AUCACCAGCUCUCCUA ....((((.(((((((.(((((((((((.....)))))).))))).))))))).)))).--.(((((......((....--.......))((((.(.(..----..).)))))))))).. ( -40.80) >DroSec_CAF1 47886 113 + 1 UAC-GGGGACUUUGAUAGAUUACAGGUGUAAAGCGCCUGCUAAUCAAUCGAAGCUCCCA--UGGUGAAAAAUCGAUAAAAAAAAACCUUCGCUGAGCGGU----AACACCAGCUCUCCAA ...-(((..(((((((.(((((((((((.....)))))).))))).)))))))..))).--.((((((...................))))))((((((.----....)).))))..... ( -40.61) >DroEre_CAF1 46377 117 + 1 UAC-GGGCACUUUGAUAGAUUACAGGUGUAAAACGCUUGCUAAUCAAUCGAAGCUGGGAGCUAGAGAAAAAUCGAUGAA--AUAACCUUCGUUGAGUGGUCGGUCAACCAAAUUCUCGUA ..(-.(((..((((((.(((((((((((.....)))))).))))).))))))))).)......(((((...((((((((--......)))))))).((((......))))..)))))... ( -35.60) >consensus UAC_GGGAACUUUGAUAGAUUACAGGUGUAAAGCGCCUGCUAAUCAAUCGAAGCUCCCA__UGGAGAAAAAUCGAUAAA__AUAACCUUCGCUGAGCGGU____AACACCAGCUCUCCUA ....((((.(((((((.(((((((((((.....)))))).))))).))))))).))))...................................((((..............))))..... (-27.53 = -27.87 + 0.34)

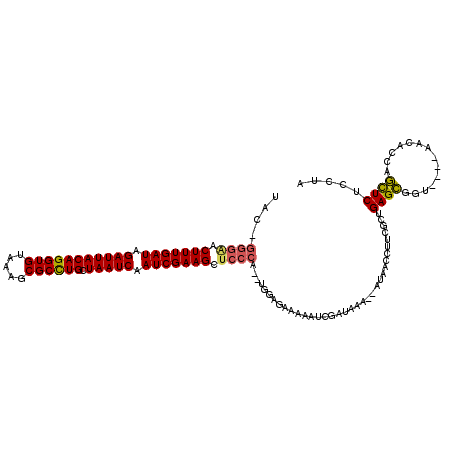
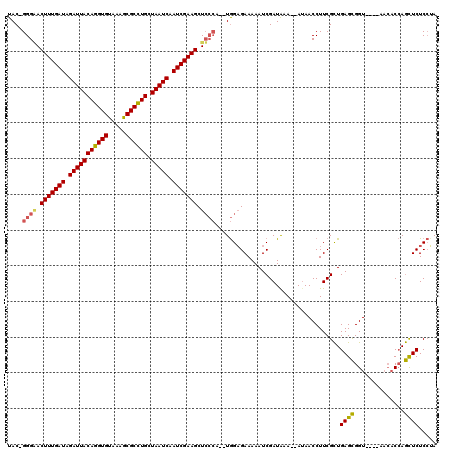
| Location | 22,289,482 – 22,289,594 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -20.66 |
| Energy contribution | -22.00 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22289482 112 - 23771897 UAGGAGAGCUGGUGAU----ACCGCUCAGCGAAGGUUAU--UUUAUCGAUUUUUCUCCA--UGGGAGCUUCGAUUGAUUAGCAGGCGCUUUACACCUGUAAUCUAUCAAAGUUCCCGGUA ..((((((((((((..----..))).))))...(((...--...)))......))))).--(((((((((.(((.(((((.((((.(.....).))))))))).))).)))))))))... ( -42.90) >DroSec_CAF1 47886 113 - 1 UUGGAGAGCUGGUGUU----ACCGCUCAGCGAAGGUUUUUUUUUAUCGAUUUUUCACCA--UGGGAGCUUCGAUUGAUUAGCAGGCGCUUUACACCUGUAAUCUAUCAAAGUCCCC-GUA ..((.((((.((....----.))))))(((....)))...................))(--((((..(((.(((.(((((.((((.(.....).))))))))).))).)))..)))-)). ( -35.20) >DroEre_CAF1 46377 117 - 1 UACGAGAAUUUGGUUGACCGACCACUCAACGAAGGUUAU--UUCAUCGAUUUUUCUCUAGCUCCCAGCUUCGAUUGAUUAGCAAGCGUUUUACACCUGUAAUCUAUCAAAGUGCCC-GUA ((((.(..((((((.((..........((((...((((.--((.(((((.........(((.....)))))))).)).))))...))))((((....)))))).))))))...).)-))) ( -21.50) >consensus UAGGAGAGCUGGUGUU____ACCGCUCAGCGAAGGUUAU__UUUAUCGAUUUUUCUCCA__UGGGAGCUUCGAUUGAUUAGCAGGCGCUUUACACCUGUAAUCUAUCAAAGUCCCC_GUA ...((((((.((.........))))))(((....)))........))...............(((.((((.(((.(((((.((((.(.....).))))))))).))).)))).))).... (-20.66 = -22.00 + 1.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:17:05 2006