
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,283,322 – 22,283,482 |
| Length | 160 |
| Max. P | 0.999177 |

| Location | 22,283,322 – 22,283,442 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -42.10 |
| Consensus MFE | -41.44 |
| Energy contribution | -41.32 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.999177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
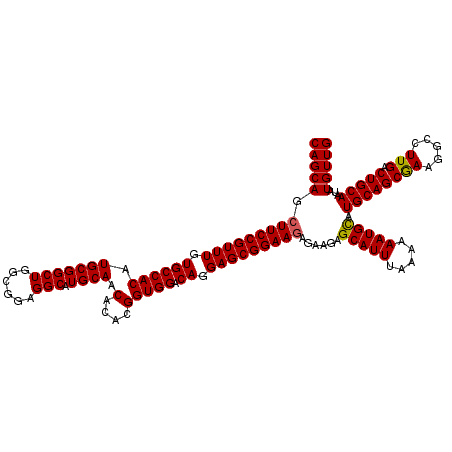
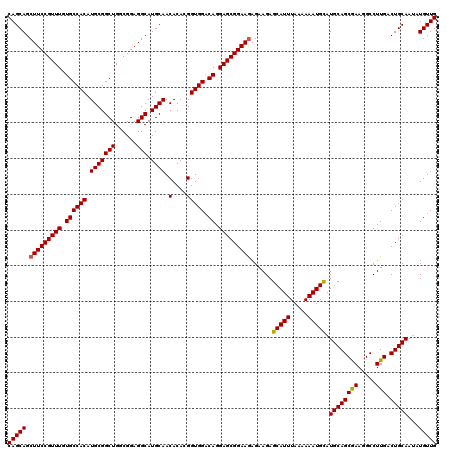
>3L_DroMel_CAF1 22283322 120 - 23771897 CAGCAGCUUCCGUUUGUGCCACAUGCGGCUGGCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAGAGAAGAGCAUUUAAAAAAUGUAUGCAGCGAAGGCCUUGACUGCAAAAUGUUG (((((.(((((((((.((((((.(((((((......))).)))).(....))))).)).)))))))))......(((((.....))))).((((((((.....))).)))))...))))) ( -40.20) >DroSec_CAF1 41692 120 - 1 CAGCAGCUUCCGUUUGUGCCACAUGCGGCUGGCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAGAGAAGAGCAUUUAAAAAAUGCAUGCAGCGAAGGCCUUGACUGCAAUAUGUUG (((((.(((((((((.((((((.(((((((......))).)))).(....))))).)).)))))))))......(((((.....))))).((((((((.....))).)))))...))))) ( -42.50) >DroSim_CAF1 33719 120 - 1 CAGCAGCUUCCGUUUGUGCCACAUGCGGCUGGCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAGAGAAGAGCAUUUAAAAAAUGCAUGCAGCGAAGGCCUUGACUGCAAUAUGUUG (((((.(((((((((.((((((.(((((((......))).)))).(....))))).)).)))))))))......(((((.....))))).((((((((.....))).)))))...))))) ( -42.50) >DroEre_CAF1 40263 120 - 1 CAGCAGCUUCCGUUUGUGCCACAUGCGGCUGUCGGAGGCAUGCAACACACGGUGGGCAGGAGCGGAAGAGGAGAGCAUUUAAAAAAUGCAUGCAGCAAAGGCCUUGACUGCAAUAUGUUG (((((.(((((((((.((((...(((((((......))).)))).(((...))))))).)))))))))......(((((.....))))).((((((((.....))).)))))...))))) ( -45.00) >DroYak_CAF1 6332 120 - 1 CAGCAGCUUCCGUUUGUGCCACAUGCGGCUGUCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAAAGGAGAGCAUUUAAAAAAUGCAUGCAGCGAAGGCCUUGACUGCAAUAUGUUG (((((..((((((((.((((((.((..(((((.....))).))..))....)))).)).)))))))).......(((((.....))))).((((((((.....))).)))))...))))) ( -40.30) >consensus CAGCAGCUUCCGUUUGUGCCACAUGCGGCUGGCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAGAGAAGAGCAUUUAAAAAAUGCAUGCAGCGAAGGCCUUGACUGCAAUAUGUUG (((((.(((((((((.((((((.(((((((......))).)))).(....))))).)).)))))))))......(((((.....))))).((((((((.....))).)))))...))))) (-41.44 = -41.32 + -0.12)

| Location | 22,283,362 – 22,283,482 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -43.80 |
| Consensus MFE | -43.26 |
| Energy contribution | -42.94 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
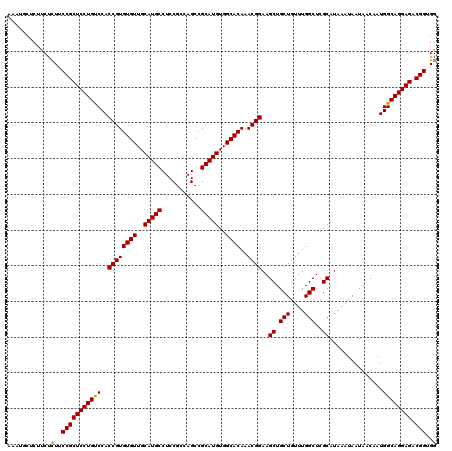
>3L_DroMel_CAF1 22283362 120 + 23771897 AAAUGCUCUUCUCUUCCGCUCCUGUCCACCGUGUGUUGCAUGCCUCCGCCAGCCGCAUGUGGCACAAACGGAAGCUGCUGUUUGGCUCGCAUAAAUAAUAACAAUGGCAGGAGACGGUGG ............((.(((((((((.(((((((((((..(((((...........)))))..))))..))))..((.(((....)))..))..............))))))))).))).)) ( -43.20) >DroSec_CAF1 41732 120 + 1 AAAUGCUCUUCUCUUCCGCUCCUGUCCACCGUGUGUUGCAUGCCUCCGCCAGCCGCAUGUGGCACAAACGGAAGCUGCUGUUUGGCUCGCAUAAAUAAUAACAAUGGCAGGAGACGGUGG ............((.(((((((((.(((((((((((..(((((...........)))))..))))..))))..((.(((....)))..))..............))))))))).))).)) ( -43.20) >DroSim_CAF1 33759 120 + 1 AAAUGCUCUUCUCUUCCGCUCCUGUCCACCGUGUGUUGCAUGCCUCCGCCAGCCGCAUGUGGCACAAACGGAAGCUGCUGUUUGGCUCGCAUAAAUAAUAACAAUGGCAGGAGACGGUGG ............((.(((((((((.(((((((((((..(((((...........)))))..))))..))))..((.(((....)))..))..............))))))))).))).)) ( -43.20) >DroEre_CAF1 40303 120 + 1 AAAUGCUCUCCUCUUCCGCUCCUGCCCACCGUGUGUUGCAUGCCUCCGACAGCCGCAUGUGGCACAAACGGAAGCUGCUGUUUGGCUCGCAUAAAUAAUAACAAUGGCAGGAGACGGUGG .........((....(((((((((((..((((((((..(((((...........)))))..))))..))))..((.(((....)))..))...............)))))))).))).)) ( -45.80) >DroYak_CAF1 6372 120 + 1 AAAUGCUCUCCUUUUCCGCUCCUGUCCACCGUGUGUUGCAUGCCUCCGACAGCCGCAUGUGGCACAAACGGAAGCUGCUGUUUGGCUCGCAUAAAUAAUAACAAUGGCAGGAGACGGUGG .........((....(((((((((.(((((((((((..(((((...........)))))..))))..))))..((.(((....)))..))..............))))))))).))).)) ( -43.60) >consensus AAAUGCUCUUCUCUUCCGCUCCUGUCCACCGUGUGUUGCAUGCCUCCGCCAGCCGCAUGUGGCACAAACGGAAGCUGCUGUUUGGCUCGCAUAAAUAAUAACAAUGGCAGGAGACGGUGG ............(..(((((((((((..((((((((..(((((...........)))))..))))..))))..((.(((....)))..))...............)))))))).)))..) (-43.26 = -42.94 + -0.32)


| Location | 22,283,362 – 22,283,482 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -45.46 |
| Consensus MFE | -44.82 |
| Energy contribution | -45.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
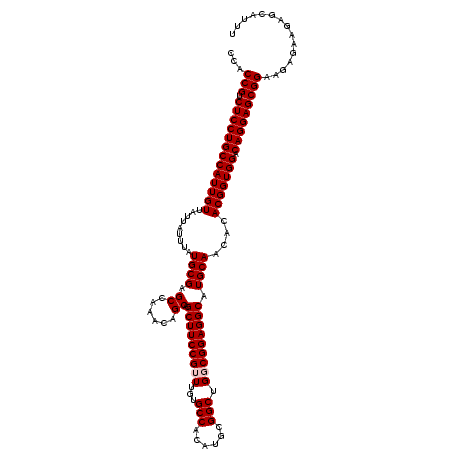
>3L_DroMel_CAF1 22283362 120 - 23771897 CCACCGUCUCCUGCCAUUGUUAUUAUUUAUGCGAGCCAAACAGCAGCUUCCGUUUGUGCCACAUGCGGCUGGCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAGAGAAGAGCAUUU ...(((.(((((((((((((.........((((.((......)).(((((((((...(((......))).))))))))).))))....))))))).)))))))))............... ( -46.42) >DroSec_CAF1 41732 120 - 1 CCACCGUCUCCUGCCAUUGUUAUUAUUUAUGCGAGCCAAACAGCAGCUUCCGUUUGUGCCACAUGCGGCUGGCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAGAGAAGAGCAUUU ...(((.(((((((((((((.........((((.((......)).(((((((((...(((......))).))))))))).))))....))))))).)))))))))............... ( -46.42) >DroSim_CAF1 33759 120 - 1 CCACCGUCUCCUGCCAUUGUUAUUAUUUAUGCGAGCCAAACAGCAGCUUCCGUUUGUGCCACAUGCGGCUGGCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAGAGAAGAGCAUUU ...(((.(((((((((((((.........((((.((......)).(((((((((...(((......))).))))))))).))))....))))))).)))))))))............... ( -46.42) >DroEre_CAF1 40303 120 - 1 CCACCGUCUCCUGCCAUUGUUAUUAUUUAUGCGAGCCAAACAGCAGCUUCCGUUUGUGCCACAUGCGGCUGUCGGAGGCAUGCAACACACGGUGGGCAGGAGCGGAAGAGGAGAGCAUUU ...(((.((((((((..............((((.((......)).(((((((.....(((......)))...))))))).)))).(((...))))))))))))))............... ( -44.50) >DroYak_CAF1 6372 120 - 1 CCACCGUCUCCUGCCAUUGUUAUUAUUUAUGCGAGCCAAACAGCAGCUUCCGUUUGUGCCACAUGCGGCUGUCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAAAGGAGAGCAUUU ...(((.(((((((((((((.........((((.((......)).(((((((.....(((......)))...))))))).))))....))))))).)))))))))............... ( -43.52) >consensus CCACCGUCUCCUGCCAUUGUUAUUAUUUAUGCGAGCCAAACAGCAGCUUCCGUUUGUGCCACAUGCGGCUGGCGGAGGCAUGCAACACACGGUGGACAGGAGCGGAAGAGAAGAGCAUUU ...(((.(((((((((((((.........((((.((......)).(((((((((...(((......))).))))))))).))))....))))))).)))))))))............... (-44.82 = -45.22 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:55 2006