
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,282,233 – 22,282,393 |
| Length | 160 |
| Max. P | 0.707299 |

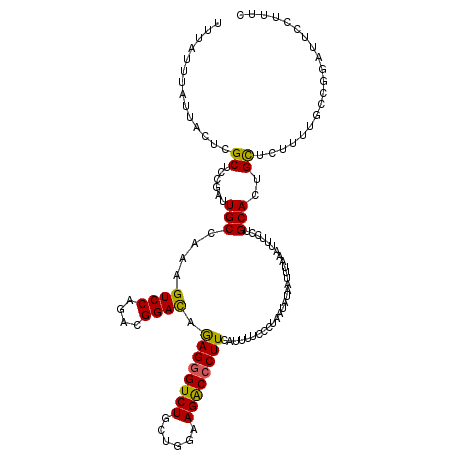
| Location | 22,282,233 – 22,282,353 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.25 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -18.86 |
| Energy contribution | -18.34 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22282233 120 + 23771897 UUUAUUUAUUACUCGCUCCGAUUGCAAAAGUCCAGACGGACAGAGGGUCUGCUGGAAGACCCUUCAUUUUUCCUAAUAUAAUUUAAAUUUCCUGCACUGCUCUUUCUCCGGAUUCCAUUC ................((((...(((...((((....)))).((((((((......)))))))).................................)))........))))........ ( -23.50) >DroSec_CAF1 40605 120 + 1 UUUAUUUGUUACUCGCUCCGAUUGCCAAAGUCCAGACGGACAGAGGGUCUGCUGGCAGACCCUUCAUUUUCCCUAAUAUAAUUUAAAUUUCCUGCACUGCUCUUUUGCCGGAUUCCUUUC ................((((..(((....((((....)))).(((((((((....))))))))).............................)))..((......))))))........ ( -29.00) >DroSim_CAF1 32614 120 + 1 UUUAUUUAUUACUCGCUCCGAUUGCCAAAGUCCAGACGGACAGAGGGUCUGCUGGAAGACCCUUCAUUUUCCCUAAUAUAAUUUAAAUUUCCUGCACUGCUCUUUUGCCGGAUUCCUUUC ................((((..(((....((((....)))).((((((((......)))))))).............................)))..((......))))))........ ( -26.10) >DroEre_CAF1 39267 120 + 1 UUUAUUUAUUACUCGCUCCGAUUGCCAAAGUCCAAACGGACAGAGGGUCUGCUGGCAGGCCCUUCAUCUUUCCUAAUAUAAUUUAAAUUUCCAGCACUGCUCUUUUGUCGGAUUCCUUUC ................(((((((((....((((....)))).(((((((((....))))))))).............................)))..........))))))........ ( -28.70) >DroYak_CAF1 5295 106 + 1 UUUAUUUAUUACUCGCUCCGAUUGCAAAAGUCCAAACGGAUAAAGUGUCUGCAGGAAGGCCCUUCAUCUUCCCUAAUAUAAUUUAAAUUUCCUGCACUGUUCAUCU-------------- .........((((...((((.(((........))).))))...))))..(((((((((.............................)))))))))..........-------------- ( -17.55) >consensus UUUAUUUAUUACUCGCUCCGAUUGCCAAAGUCCAGACGGACAGAGGGUCUGCUGGAAGACCCUUCAUUUUCCCUAAUAUAAUUUAAAUUUCCUGCACUGCUCUUUUGCCGGAUUCCUUUC ..............((......(((....((((....)))).((((((((......)))))))).............................)))..)).................... (-18.86 = -18.34 + -0.52)


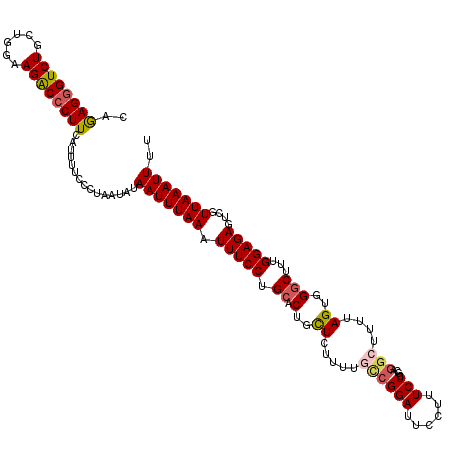
| Location | 22,282,273 – 22,282,393 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.78 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22282273 120 + 23771897 CAGAGGGUCUGCUGGAAGACCCUUCAUUUUUCCUAAUAUAAUUUAAAUUUCCUGCACUGCUCUUUCUCCGGAUUCCAUUCUCGCUGGCUUUUAGUGGGCUUUGGAGAGUCGUUAAAUUUU ..((((((((......))))))))...............(((((((.(((((.((.(..((......((((............)))).....))..)))...)))))....))))))).. ( -29.20) >DroSec_CAF1 40645 120 + 1 CAGAGGGUCUGCUGGCAGACCCUUCAUUUUCCCUAAUAUAAUUUAAAUUUCCUGCACUGCUCUUUUGCCGGAUUCCUUUCUCGCAGGCUUUUAGUGGUCUUUGGAGAGUCGUUAAAUUUU ..(((((((((....)))))))))...............(((((((.(((((.(.((..((..(((((.(((......))).))))).....))..)))...)))))....))))))).. ( -33.00) >DroSim_CAF1 32654 120 + 1 CAGAGGGUCUGCUGGAAGACCCUUCAUUUUCCCUAAUAUAAUUUAAAUUUCCUGCACUGCUCUUUUGCCGGAUUCCUUUCUCGCAGGCUUUUAGUGGGCUUUGGAGAGUCGUUAAAUUUU ..((((((((......))))))))...............(((((((.(((((.((.(..((..(((((.(((......))).))))).....))..)))...)))))....))))))).. ( -32.20) >DroEre_CAF1 39307 120 + 1 CAGAGGGUCUGCUGGCAGGCCCUUCAUCUUUCCUAAUAUAAUUUAAAUUUCCAGCACUGCUCUUUUGUCGGAUUCCUUUCUCUUUAGCUUUUAGUGGGCUUUGGAGAGUCGUUAAAUUUU ..(((((((((....)))))))))...............(((((((...(((.(((.........))).))).....((((((..(((((.....)))))..))))))...))))))).. ( -32.00) >DroYak_CAF1 5335 100 + 1 UAAAGUGUCUGCAGGAAGGCCCUUCAUCUUCCCUAAUAUAAUUUAAAUUUCCUGCACUGUUCAUCU--------------------UCUUUUAGUGGGCAUUGGAGAGUCGUUAAAUUUU .............((((((.......)))))).......(((((((.(((((.....((((((.((--------------------......))))))))..)))))....))))))).. ( -17.20) >consensus CAGAGGGUCUGCUGGAAGACCCUUCAUUUUCCCUAAUAUAAUUUAAAUUUCCUGCACUGCUCUUUUGCCGGAUUCCUUUCUCGCAGGCUUUUAGUGGGCUUUGGAGAGUCGUUAAAUUUU ..((((((((......))))))))...............(((((((.(((((.((.(..((.....((((((......)))....)))....))..)))...)))))....))))))).. (-22.06 = -22.78 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:48 2006