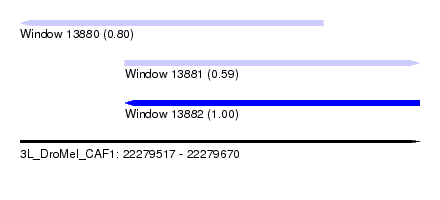
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,279,517 – 22,279,670 |
| Length | 153 |
| Max. P | 0.996543 |

| Location | 22,279,517 – 22,279,633 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -31.58 |
| Energy contribution | -33.20 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22279517 116 - 23771897 AGAUGCAGAAGGAUGGAUUUGCCCGUCAGUCCCAAGGGCUGUGCGGAAGAACAACA----UCGCAGAAAACAGGCUCGAAUCAAUGUUUAAUCCUCGCCAGCAGCCGGAAACCUGGCCGC ..........((..(((((...((((((((((...)))))).))))..(((((...----(((.((........))))).....))))))))))...)).((.(((((....))))).)) ( -33.50) >DroSec_CAF1 37897 116 - 1 AGAUGCAGAAGGAUGGAUUUGCCCGUCAGCCCCAAGGGCUGUGCGGAAGCACAACA----UCGCAGAAAACAGGCUUGAAUCAAAGUUUGAUCCUCGCCAGCAGCCGGAAACCUGGCCGC ....((.((.((((...(((((.....(((((...)))))((((....))))....----..)))))...(((((((......)))))))))))))))..((.(((((....))))).)) ( -42.30) >DroSim_CAF1 29493 116 - 1 AGAUGCAGAAGGAUGGAUUUGCCCGUCAGCCCCAAGGGCUGUGCGGAAGCACAACA----UCGCAGAAAACAGGCUCGAAUCAAAGUUUGAUCCUCGCCAGCAGCCGGAAACCUGGCCGC .((((......(((((......)))))(((((...)))))((((....))))..))----))..........(((..(.((((.....)))).)..))).((.(((((....))))).)) ( -41.30) >DroEre_CAF1 36584 116 - 1 AGAUGCAGAAGGAUGGAGUUACCCGUCAGCCCCAAGGGCUCUGCGGAAGCACAACA----UGGCAGAAAACAGGCUCGAAUCAAAGUUUGAUCCUCGCCAGCAGCGAGAAACCUGGCCGC ....(((((..(((((......))))).((((...)))))))))((..((......----..))......((((.((((((....))))))..(((((.....)))))...)))).)).. ( -37.50) >DroYak_CAF1 2665 106 - 1 AGAUGCAGCAGGAUGGAUUUACCCGUCAGCCCCAAGGGCUGUGCGGAAGCACAACAUACAUAGCAGAAAACAGGCUCGAAUCAAAGUUUGAUCCUCGCCAGCAGCA-------------- .(.(((.((..(((((......)))))(((((...)))))((((....))))..........))........(((..(.((((.....)))).)..))).))).).-------------- ( -34.40) >consensus AGAUGCAGAAGGAUGGAUUUGCCCGUCAGCCCCAAGGGCUGUGCGGAAGCACAACA____UCGCAGAAAACAGGCUCGAAUCAAAGUUUGAUCCUCGCCAGCAGCCGGAAACCUGGCCGC ...(((.....(((((......)))))(((((...)))))((((....))))..........))).......(((..(.((((.....)))).)..))).((.(((((....))))).)) (-31.58 = -33.20 + 1.62)

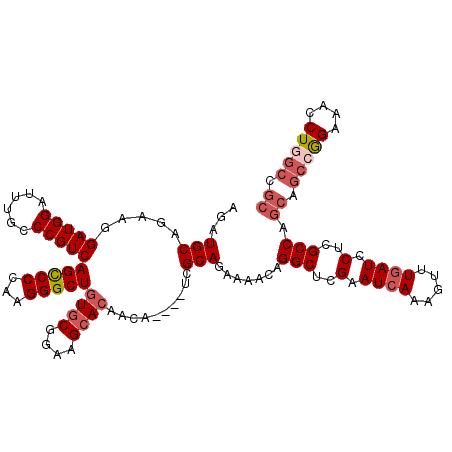
| Location | 22,279,557 – 22,279,670 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -42.32 |
| Consensus MFE | -30.76 |
| Energy contribution | -32.72 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22279557 113 + 23771897 UUCGAGCCUGUUUUCUGCGA----UGUUGUUCUUCCGCACAGCCCUUGGGACUGACGGGCAAAUCCAUCCUUCUGCAUCUGCAGCGUGC---UGUAGCGUACACUGCGGCUGUGUUAAUC .....((((((....((((.----...........))))(((((....)).)))))))))..............(((.((((((.((((---......)))).)))))).)))....... ( -35.60) >DroSec_CAF1 37937 116 + 1 UUCAAGCCUGUUUUCUGCGA----UGUUGUGCUUCCGCACAGCCCUUGGGGCUGACGGGCAAAUCCAUCCUUCUGCAUCUGCAGCGUGCUGCUGUAGCGUACGCUGCGGCUGGGUUAAUC .....(((((((..((.(((----.(((((((....)))))))..))).))..))))))).((((((.((..........(((((((((.((....))))))))))))).)))))).... ( -50.20) >DroSim_CAF1 29533 116 + 1 UUCGAGCCUGUUUUCUGCGA----UGUUGUGCUUCCGCACAGCCCUUGGGGCUGACGGGCAAAUCCAUCCUUCUGCAUCUGCAGCGUGCUGCUGUAGCGUACACUGCGGCUGGGUUAAUC .....(((((((..((.(((----.(((((((....)))))))..))).))..))))))).((((((.....((((....))))......(((((((......))))))))))))).... ( -45.00) >DroEre_CAF1 36624 113 + 1 UUCGAGCCUGUUUUCUGCCA----UGUUGUGCUUCCGCAGAGCCCUUGGGGCUGACGGGUAACUCCAUCCUUCUGCAUCUGCAGCGUGC---UGUAGCAUACUCUGCGGCGGGGUUAAUC .....(((((((..((.(((----.(((.(((....))).)))...)))))..)))))))((((((......((((....))))...((---(((((......))))))))))))).... ( -40.90) >DroYak_CAF1 2691 117 + 1 UUCGAGCCUGUUUUCUGCUAUGUAUGUUGUGCUUCCGCACAGCCCUUGGGGCUGACGGGUAAAUCCAUCCUGCUGCAUCUGCAGCCAGC---UGUACCAUACUCUGCGGCGGUGUUAAUC .....(((((((....(((......(((((((....)))))))......))).))))))).....((((..(((((....)))))..((---((((........))))))))))...... ( -39.90) >consensus UUCGAGCCUGUUUUCUGCGA____UGUUGUGCUUCCGCACAGCCCUUGGGGCUGACGGGCAAAUCCAUCCUUCUGCAUCUGCAGCGUGC___UGUAGCGUACACUGCGGCUGGGUUAAUC .....(((((((..((.(((.....(((((((....)))))))..))).))..))))))).((((((.((..((((....)))).........((((......)))))).)))))).... (-30.76 = -32.72 + 1.96)

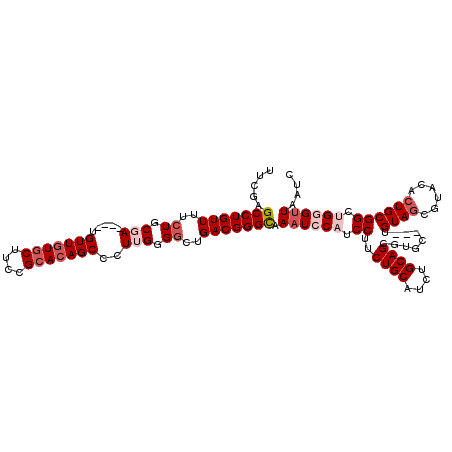
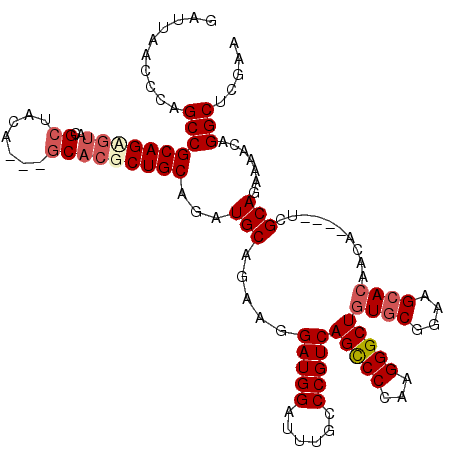
| Location | 22,279,557 – 22,279,670 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -35.50 |
| Energy contribution | -36.66 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22279557 113 - 23771897 GAUUAACACAGCCGCAGUGUACGCUACA---GCACGCUGCAGAUGCAGAAGGAUGGAUUUGCCCGUCAGUCCCAAGGGCUGUGCGGAAGAACAACA----UCGCAGAAAACAGGCUCGAA .........(((((((((((..((....---)))))))))...(((.....((((..(((..((((((((((...)))))).))))..)))...))----))))).......)))).... ( -36.80) >DroSec_CAF1 37937 116 - 1 GAUUAACCCAGCCGCAGCGUACGCUACAGCAGCACGCUGCAGAUGCAGAAGGAUGGAUUUGCCCGUCAGCCCCAAGGGCUGUGCGGAAGCACAACA----UCGCAGAAAACAGGCUUGAA .......(.(((((((((((.(((....)).).))))))).((((......(((((......)))))(((((...)))))((((....))))..))----))..........)))).).. ( -46.60) >DroSim_CAF1 29533 116 - 1 GAUUAACCCAGCCGCAGUGUACGCUACAGCAGCACGCUGCAGAUGCAGAAGGAUGGAUUUGCCCGUCAGCCCCAAGGGCUGUGCGGAAGCACAACA----UCGCAGAAAACAGGCUCGAA .......(.(((((((((((.(((....)).).))))))).((((......(((((......)))))(((((...)))))((((....))))..))----))..........)))).).. ( -44.70) >DroEre_CAF1 36624 113 - 1 GAUUAACCCCGCCGCAGAGUAUGCUACA---GCACGCUGCAGAUGCAGAAGGAUGGAGUUACCCGUCAGCCCCAAGGGCUCUGCGGAAGCACAACA----UGGCAGAAAACAGGCUCGAA ..........(((((((....(((....---)))..))))...((((((..(((((......))))).((((...)))))))((....))......----..))).......)))..... ( -34.80) >DroYak_CAF1 2691 117 - 1 GAUUAACACCGCCGCAGAGUAUGGUACA---GCUGGCUGCAGAUGCAGCAGGAUGGAUUUACCCGUCAGCCCCAAGGGCUGUGCGGAAGCACAACAUACAUAGCAGAAAACAGGCUCGAA ..........(((((...(((((.....---.(((.((((....)))))))(((((......)))))(((((...)))))((((....))))..)))))...)).(....).)))..... ( -40.70) >consensus GAUUAACCCAGCCGCAGAGUACGCUACA___GCACGCUGCAGAUGCAGAAGGAUGGAUUUGCCCGUCAGCCCCAAGGGCUGUGCGGAAGCACAACA____UCGCAGAAAACAGGCUCGAA ..........((((((((((..((.......)))))))))...(((.....(((((......)))))(((((...)))))((((....))))..........))).......)))..... (-35.50 = -36.66 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:42 2006