
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,278,353 – 22,278,697 |
| Length | 344 |
| Max. P | 0.999975 |

| Location | 22,278,353 – 22,278,469 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -19.52 |
| Energy contribution | -21.32 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22278353 116 + 23771897 AGACAGCACUGAUGGCAGGUAGAAACAAGUAGUCCGCACUGAACGGCAUAGAAUAAAUGCAAGUGCUCAUGUGUGC----UAUGCUGCGUAUACGCAGUGUAGUACAGGGAGUGCGAUUU .....((.(((....)))))........(((.((((((((.....((((.......)))).))))).....(((((----(((((((((....)))))))))))))).))).)))..... ( -42.60) >DroSec_CAF1 36764 115 + 1 AGACAGCAAUAAUGGCAGGUAGAAACAAGUAGUCCGCACUGAACGGCAUUGAAUAAAUGCAAGUGCUCAUGUGUGC----UACACUGCGUAUACGCAAUGU-GCACAGGGAUUGCGAUUU .....((.......))............((((((((((((.....(((((.....))))).))))).....(((((----.(((.((((....)))).)))-))))).)))))))..... ( -38.50) >DroSim_CAF1 28351 115 + 1 AGACAGCAAUGAUGGCAGGUAGAAACAAGUAGUCCGCACUGAACGGCAUUGAAUAAAUGCAAGUGCUCAUGUGUGC----UACACUGCGUAUACGCAAUGU-GCACAGGGAUUGCGAUUU .....((.......))............((((((((((((.....(((((.....))))).))))).....(((((----.(((.((((....)))).)))-))))).)))))))..... ( -38.50) >DroEre_CAF1 35524 98 + 1 GGACACAAAUGAUGACUGGUAGAAGCAAG-----------------CAUUGAAUAAAUGCAAGUGCCCAUGUGUGC----UACACUGCGUAUACGCAACGU-GUACAGGGAGUGCUAUUU ((.(((......((.((......)))).(-----------------((((.....)))))..))).))....(..(----(...((..(((((((...)))-)))).)).))..)..... ( -24.30) >DroYak_CAF1 1538 119 + 1 AGACAGCAAUGAUGACAGGUAGAAACAAGUAGUCCGCACUGAACGGCAUUAAAUAAAUGCAAGUGCUCAUAUGUGCUAGCUACACUGCGUAUACGCAACGU-GUGCAGGUAGUGCGAUUU ((..((((...((((..((..............))(((((.....(((((.....))))).)))))))))...))))..)).(((((((((((((...)))-))))..))))))...... ( -32.94) >consensus AGACAGCAAUGAUGGCAGGUAGAAACAAGUAGUCCGCACUGAACGGCAUUGAAUAAAUGCAAGUGCUCAUGUGUGC____UACACUGCGUAUACGCAAUGU_GUACAGGGAGUGCGAUUU .....(((((.........................(((((.....(((((.....))))).)))))...(.(((((.....(((.((((....)))).))).))))).).)))))..... (-19.52 = -21.32 + 1.80)

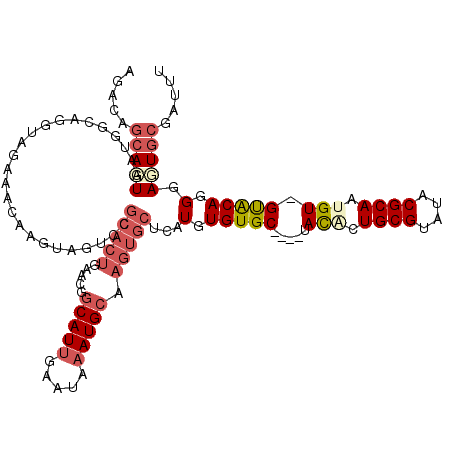
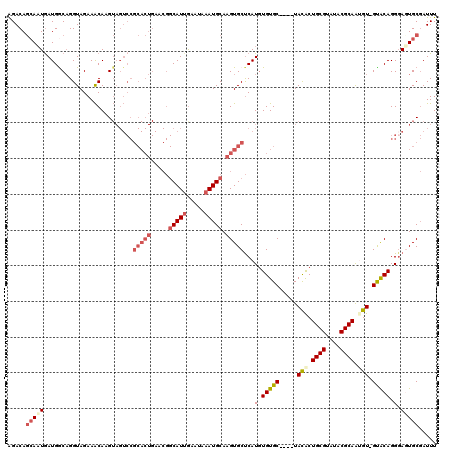
| Location | 22,278,393 – 22,278,503 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22278393 110 - 23771897 ---UUCCCCAG--U-UCCCCACCCUGCUCGUCACAACAAAAAAUCGCACUCCCUGUACUACACUGCGUAUACGCAGCAUA----GCACACAUGAGCACUUGCAUUUAUUCUAUGCCGUUC ---........--.-.........(((((((....))................(((.(((..(((((....)))))..))----).)))...)))))...((((.......))))..... ( -19.70) >DroSec_CAF1 36804 110 - 1 ---UUCCCUAG--CAUCCCCACCCUGCUCGUCACAACAAAAAAUCGCAAUCCCUGUGC-ACAUUGCGUAUACGCAGUGUA----GCACACAUGAGCACUUGCAUUUAUUCAAUGCCGUUC ---......((--(..........(((((((....))................(((((-((((((((....)))))))).----)))))...)))))...(((((.....))))).))). ( -27.80) >DroSim_CAF1 28391 113 - 1 AGCUUCCCUAG--CAUCCCCACCCUGCUCGUCACAACAAAAAAUCGCAAUCCCUGUGC-ACAUUGCGUAUACGCAGUGUA----GCACACAUGAGCACUUGCAUUUAUUCAAUGCCGUUC .(((.....))--)..........(((((((....))................(((((-((((((((....)))))))).----)))))...)))))...(((((.....)))))..... ( -30.10) >DroEre_CAF1 35553 96 - 1 -------------CCUGCCCAUCCUGCCCAUCACGACAAAAAAUAGCACUCCCUGUAC-ACGUUGCGUAUACGCAGUGUA----GCACACAUGGGCACUUGCAUUUAUUCAAUG------ -------------..(((((((..(((...............((((......))))((-((..((((....)))))))).----)))...))))))).................------ ( -21.70) >DroYak_CAF1 1578 116 - 1 ---ACCCCCAGGACUUCCCCAUUCUGCCAAUCACAACAAAAAAUCGCACUACCUGCAC-ACGUUGCGUAUACGCAGUGUAGCUAGCACAUAUGAGCACUUGCAUUUAUUUAAUGCCGUUC ---.....(((((........)))))...................(..(((.((((((-....((((....)))))))))).)))..)....((((....(((((.....))))).)))) ( -20.40) >consensus ___UUCCCCAG__CAUCCCCACCCUGCUCGUCACAACAAAAAAUCGCACUCCCUGUAC_ACAUUGCGUAUACGCAGUGUA____GCACACAUGAGCACUUGCAUUUAUUCAAUGCCGUUC ........................(((((((..............(((.....)))...((((((((....))))))))...........)))))))...(((((.....)))))..... (-17.84 = -18.08 + 0.24)

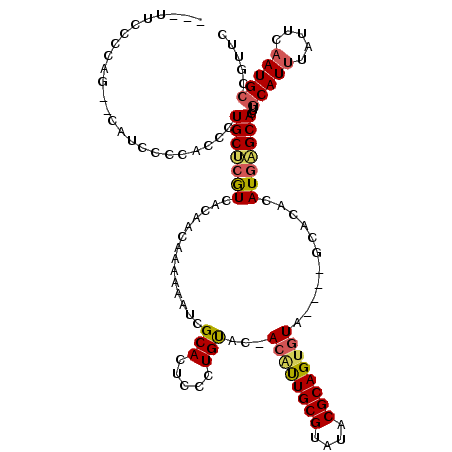
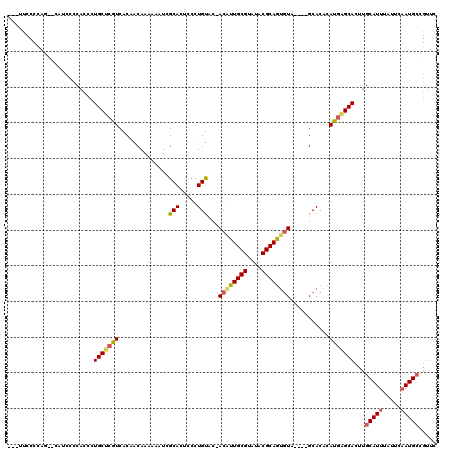
| Location | 22,278,429 – 22,278,537 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -13.47 |
| Energy contribution | -14.35 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22278429 108 - 23771897 UUAUCAACAAAAUGGCAGCAGCAGGAUCCCCAGC---------UUCCCCAG--U-UCCCCACCCUGCUCGUCACAACAAAAAAUCGCACUCCCUGUACUACACUGCGUAUACGCAGCAUA ............((((.(.((((((......(((---------(.....))--)-)......))))))))))).............................(((((....))))).... ( -20.10) >DroSec_CAF1 36840 108 - 1 UUAUCAACAAAAUGGCAGCAGCAGGACCUCCAGC---------UUCCCUAG--CAUCCCCACCCUGCUCGUCACAACAAAAAAUCGCAAUCCCUGUGC-ACAUUGCGUAUACGCAGUGUA ............(((((((((..((....)).((---------(.....))--).........))))).))))............(((.......)))-((((((((....)))))))). ( -27.70) >DroSim_CAF1 28427 117 - 1 UUAUCAACAAAAUGGCAGCAGCAGGACCUCCAGCUUCCCCAGCUUCCCUAG--CAUCCCCACCCUGCUCGUCACAACAAAAAAUCGCAAUCCCUGUGC-ACAUUGCGUAUACGCAGUGUA ............(((((((((..((....))..........(((.....))--).........))))).))))............(((.......)))-((((((((....)))))))). ( -27.70) >DroEre_CAF1 35583 90 - 1 UUAUCAACAAAAUGGCAGCAGCAU-----------------------------CCUGCCCAUCCUGCCCAUCACGACAAAAAAUAGCACUCCCUGUAC-ACGUUGCGUAUACGCAGUGUA .............(((((..((..-----------------------------...)).....))))).................(((((.(.(((((-.......))))).).))))). ( -16.30) >DroYak_CAF1 1618 110 - 1 UUAUCAACAAAAUGGCAGCAGCAGAACCCCCAGG---------ACCCCCAGGACUUCCCCAUUCUGCCAAUCACAACAAAAAAUCGCACUACCUGCAC-ACGUUGCGUAUACGCAGUGUA ............((((((.....(((..((..((---------....)).))..)))......))))))................(((.....)))((-((..((((....)))))))). ( -22.70) >consensus UUAUCAACAAAAUGGCAGCAGCAGGACCCCCAGC_________UUCCCCAG__CAUCCCCACCCUGCUCGUCACAACAAAAAAUCGCACUCCCUGUAC_ACAUUGCGUAUACGCAGUGUA ............((((...((((((.....................................)))))).))))............(((.....)))...((((((((....)))))))). (-13.47 = -14.35 + 0.88)

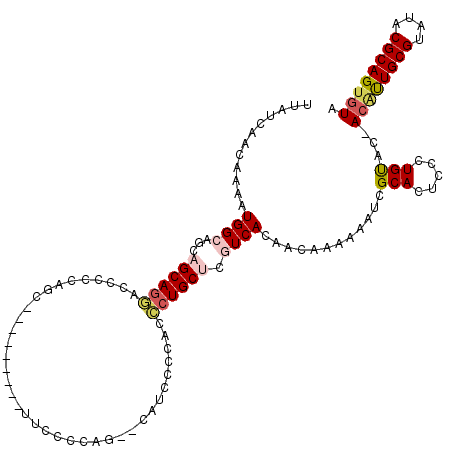
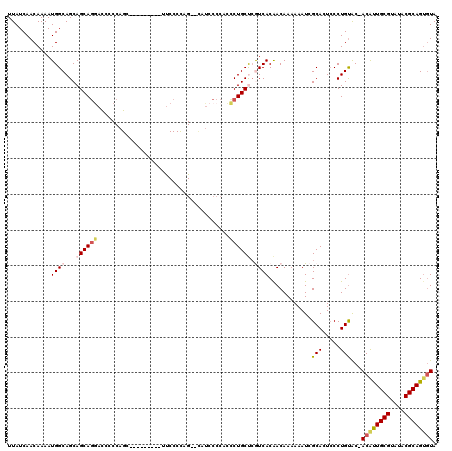
| Location | 22,278,469 – 22,278,577 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22278469 108 - 23771897 UACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGGAUCCCCAGC---------UUCCCCAG--U-UCCCCACCCUGCUCGUCACAACAAA ...........(((.......)))(((.(((((((((((((............)))))))(((((......(((---------(.....))--)-)......)))))..))))))))).. ( -26.10) >DroSec_CAF1 36879 109 - 1 UACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGGACCUCCAGC---------UUCCCUAG--CAUCCCCACCCUGCUCGUCACAACAAA ...........(((.......)))(((.(((((((((((((............)))))))(((((.......((---------(.....))--)........)))))..))))))))).. ( -27.06) >DroSim_CAF1 28466 118 - 1 UACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGGACCUCCAGCUUCCCCAGCUUCCCUAG--CAUCCCCACCCUGCUCGUCACAACAAA ..........(((......(((((((.((((.(.(((((.....)))))...).)))))))))))......((((.....))))......)--))......................... ( -26.40) >DroEre_CAF1 35622 91 - 1 UGCUUCCCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAU-----------------------------CCUGCCCAUCCUGCCCAUCACGACAAA (((........)))..........((((.((((((((((((............)))))))))..-----------------------------...((.......))....))))))).. ( -22.00) >DroYak_CAF1 1657 111 - 1 UACUUCCCUUUGCAUCUACCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGAACCCCCAGG---------ACCCCCAGGACUUCCCCAUUCUGCCAAUCACAACAAA ...........(((.......)))(((.(((((((((((((............)))))))))((((......((---------....)).((......)).))))......))))))).. ( -24.20) >consensus UACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGGACCCCCAGC_________UUCCCCAG__CAUCCCCACCCUGCUCGUCACAACAAA ...........(((.......)))(((.(((((((((((((............)))))))(((((.....................................)))))..))))))))).. (-19.17 = -19.57 + 0.40)

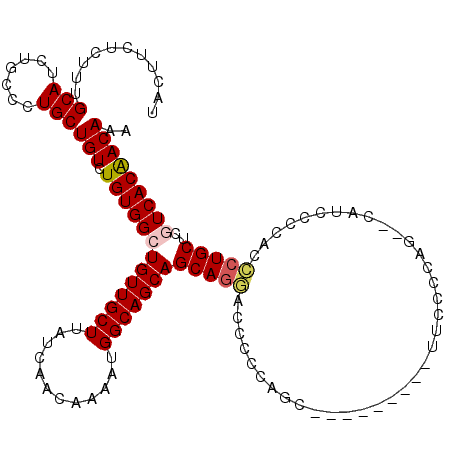
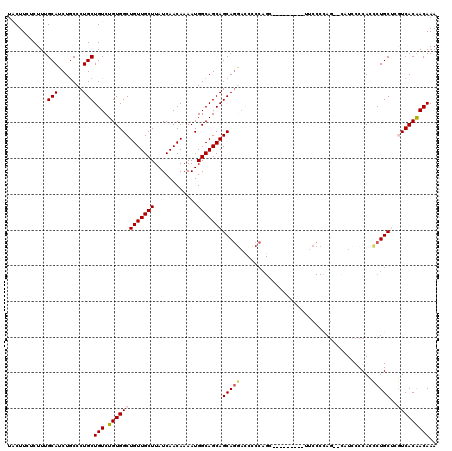
| Location | 22,278,503 – 22,278,617 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.36 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22278503 114 + 23771897 ------GCUGGGGAUCCUGCUGCUGCCAUUUUGUUGAUAAGCAACAGCCACAGACAGCAGGGCAGAUGCAAAGAGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUA ------((....(.(((((((((((.....((((((.....))))))...))).)))))))))....)).........(((((((..((((.((((........)))).))))))))))) ( -33.70) >DroSec_CAF1 36914 114 + 1 ------GCUGGAGGUCCUGCUGCUGCCAUUUUGUUGAUAAGCAACAGCCACAGACAGCAGGGCAGAUGCAAAGAGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUA ------((.....((((((((((((.....((((((.....))))))...))).)))))))))....)).........(((((((..((((.((((........)))).))))))))))) ( -36.90) >DroSim_CAF1 28504 120 + 1 GGGGAAGCUGGAGGUCCUGCUGCUGCCAUUUUGUUGAUAAGCAACAGCCACAGACAGCAGGGCAGAUGCAAAGAGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUA ......((.....((((((((((((.....((((((.....))))))...))).)))))))))....)).........(((((((..((((.((((........)))).))))))))))) ( -37.10) >DroEre_CAF1 35649 104 + 1 ----------------AUGCUGCUGCCAUUUUGUUGAUAAGCAACAGCCACAGACAGCAGGGCAGAUGCAAAGGGAAGCAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUA ----------------.(((..(((((...(((((.....((....))....)))))...)))))..))).........((((((..((((.((((........)))).)))))))))). ( -26.90) >DroYak_CAF1 1694 114 + 1 ------CCUGGGGGUUCUGCUGCUGCCAUUUUGUUGAUAAGCAACAGCCACAGACAGCAGGGUAGAUGCAAAGGGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGUUUUGUUUA ------(((..(.((((((((((((.....((((((.....))))))...))).))))))....))).)..)))....((((((((..(((.((((........)))).))))))))))) ( -29.80) >consensus ______GCUGGAGGUCCUGCUGCUGCCAUUUUGUUGAUAAGCAACAGCCACAGACAGCAGGGCAGAUGCAAAGAGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUA ...............((((((((((.....((((((.....))))))...))).)))))))((....)).........(((((((..((((.((((........)))).))))))))))) (-28.20 = -28.36 + 0.16)


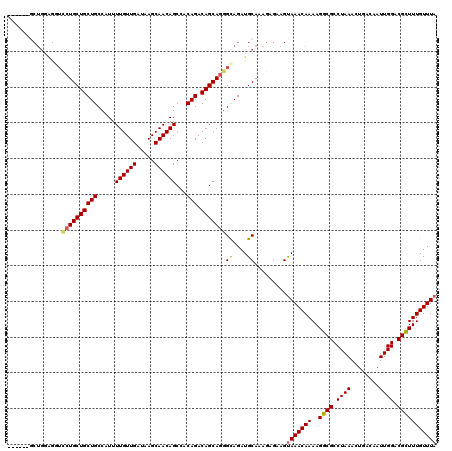
| Location | 22,278,503 – 22,278,617 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -28.32 |
| Energy contribution | -28.96 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22278503 114 - 23771897 UAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGGAUCCCCAGC------ (((((((((((((.((........)).)))))..)))))))).........((....))(((((((.((((.(.(((((.....)))))...).))))))))))).........------ ( -32.70) >DroSec_CAF1 36914 114 - 1 UAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGGACCUCCAGC------ (((((((((((((.((........)).)))))..)))))))).........((....))(((((((.((((.(.(((((.....)))))...).))))))))))).........------ ( -32.70) >DroSim_CAF1 28504 120 - 1 UAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGGACCUCCAGCUUCCCC (((((((((((((.((........)).)))))..)))))))).........((....))(((((((.((((.(.(((((.....)))))...).)))))))))))............... ( -32.70) >DroEre_CAF1 35649 104 - 1 UAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUGCUUCCCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAU---------------- .((((((((((((.((........)).)))))..)))))))((........)).......((((((((......(((((.....)))))....))))))))...---------------- ( -26.80) >DroYak_CAF1 1694 114 - 1 UAAACAAAACGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCCCUUUGCAUCUACCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGAACCCCCAGG------ (((((((((((((.((........)).))))..))))))))).................((((...(((...(((((((((............)))))))))))).....))))------ ( -26.60) >consensus UAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGCUUAUCAACAAAAUGGCAGCAGCAGGACCCCCAGC______ (((((((((((((.((........)).)))))..)))))))).................(((((((.((((.(.(((((.....)))))...).)))))))))))............... (-28.32 = -28.96 + 0.64)

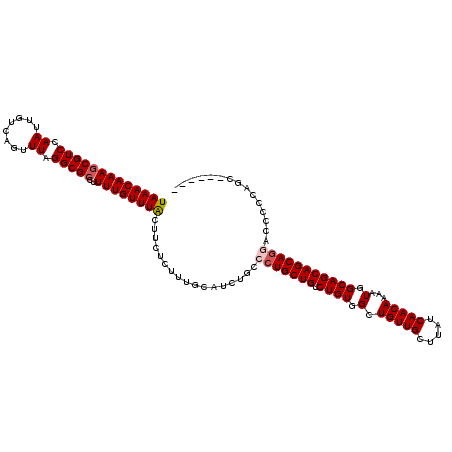

| Location | 22,278,537 – 22,278,657 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -42.22 |
| Consensus MFE | -41.62 |
| Energy contribution | -41.30 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.13 |
| SVM RNA-class probability | 0.999975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22278537 120 + 23771897 GCAACAGCCACAGACAGCAGGGCAGAUGCAAAGAGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUAUGGCGUGUCUGUGUAUUAGGGCCCUGGGUGUGUGAUUGCA ((((....((((.((..((((((.((((((.(((.(.((((((((..((((.((((........)))).))))))))))))....).))).))))))...)))))).)).)))).)))). ( -45.00) >DroSec_CAF1 36948 120 + 1 GCAACAGCCACAGACAGCAGGGCAGAUGCAAAGAGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUAUGGCGUGUCUGUGUAUUAGAGCCCUGGGUGUGUGAUUGCA ((((....((((.((..((((((.((((((.(((.(.((((((((..((((.((((........)))).))))))))))))....).))).))))))...)))))).)).)))).)))). ( -43.90) >DroSim_CAF1 28544 120 + 1 GCAACAGCCACAGACAGCAGGGCAGAUGCAAAGAGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUAUGGCGUGUCUGUGUAUUAGAGCCCUGGGUGUGUGAUUGCA ((((....((((.((..((((((.((((((.(((.(.((((((((..((((.((((........)))).))))))))))))....).))).))))))...)))))).)).)))).)))). ( -43.90) >DroEre_CAF1 35673 120 + 1 GCAACAGCCACAGACAGCAGGGCAGAUGCAAAGGGAAGCAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUAUGGCGUGUCUGUGUAUUAGAGCCCUGGGUGUGUGAUUGCA ((((....((((.((..((((((.((((((.(((.(.((......((((((.((((........)))).)))))).......)).).))).))))))...)))))).)).)))).)))). ( -41.62) >DroYak_CAF1 1728 120 + 1 GCAACAGCCACAGACAGCAGGGUAGAUGCAAAGGGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGUUUUGUUUAUGGCGUUUCUGUGUAUUAGGACCCAGGGUGUGUGAUUGCA ((((....((((.((....((((.((((((.((..(.(((((((((..(((.((((........)))).))))))))))))....)..)).))))))...))))...)).)))).)))). ( -36.70) >consensus GCAACAGCCACAGACAGCAGGGCAGAUGCAAAGAGAAGUAAACAAAAGGCGCCUAAACUGACAAUUGGACGCUUUGUUUAUGGCGUGUCUGUGUAUUAGAGCCCUGGGUGUGUGAUUGCA ((((....((((.((..((((((.((((((.(((.(.((((((((..((((.((((........)))).))))))))))))....).))).))))))...)))))).)).)))).)))). (-41.62 = -41.30 + -0.32)


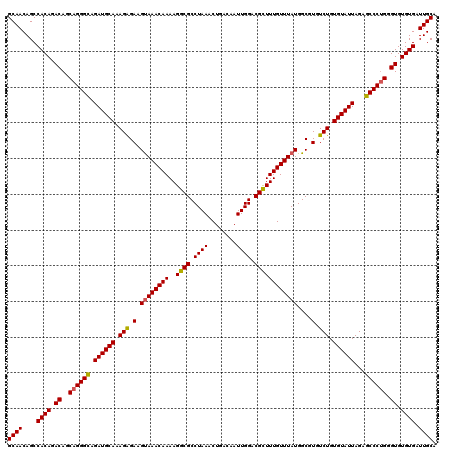
| Location | 22,278,537 – 22,278,657 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -29.22 |
| Energy contribution | -28.78 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22278537 120 - 23771897 UGCAAUCACACACCCAGGGCCCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGC .(((..((((.((.((((((....((.((.(((.......(((((((((((((.((........)).)))))..))))))))....))).)).))..))))))..)).))))..)))... ( -30.80) >DroSec_CAF1 36948 120 - 1 UGCAAUCACACACCCAGGGCUCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGC .(((..((((.((.((((((....((.((.(((.......(((((((((((((.((........)).)))))..))))))))....))).)).))..))))))..)).))))..)))... ( -31.50) >DroSim_CAF1 28544 120 - 1 UGCAAUCACACACCCAGGGCUCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGC .(((..((((.((.((((((....((.((.(((.......(((((((((((((.((........)).)))))..))))))))....))).)).))..))))))..)).))))..)))... ( -31.50) >DroEre_CAF1 35673 120 - 1 UGCAAUCACACACCCAGGGCUCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUGCUUCCCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGC .(((..((((.((.((((((........(((((..((((.(((((...(((......)))..)))))))))...))))).(((........)))...))))))..)).))))..)))... ( -35.30) >DroYak_CAF1 1728 120 - 1 UGCAAUCACACACCCUGGGUCCUAAUACACAGAAACGCCAUAAACAAAACGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCCCUUUGCAUCUACCCUGCUGUCUGUGGCUGUUGC .(((..((((.((...(((...(((...((((((.((((.(((((...(((......)))..)))))))))..)))))))))...)))...(((.......))).)).))))..)))... ( -26.80) >consensus UGCAAUCACACACCCAGGGCUCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUUUACUUCUCUUUGCAUCUGCCCUGCUGUCUGUGGCUGUUGC .(((..((((.((.((((((.....((.(((((..((((.(((((...(((......)))..)))))))))...))))).))(........).....))))))..)).))))..)))... (-29.22 = -28.78 + -0.44)

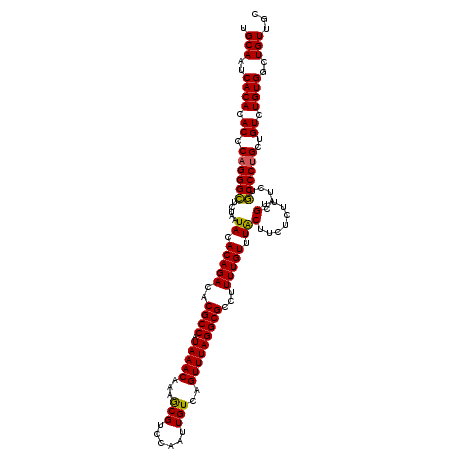
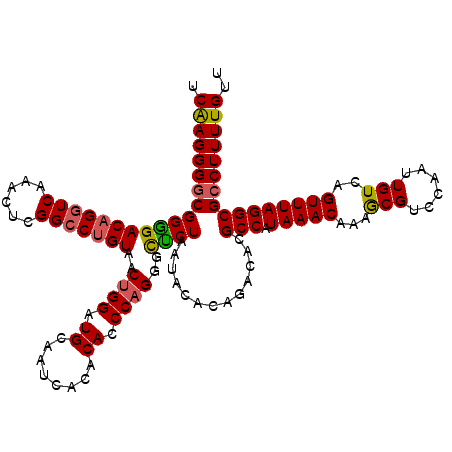
| Location | 22,278,577 – 22,278,697 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -35.62 |
| Energy contribution | -35.54 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22278577 120 - 23771897 UCAAGGGCCGGGGACAGGUCAAACUCGGCCUGUAACUGGAUGCAAUCACACACCCAGGGCCCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUU ...((((((....(((((((......)))))))..((((.((........)).)))))))))).....(((((..((((.(((((...(((......)))..)))))))))...))))). ( -41.60) >DroSec_CAF1 36988 120 - 1 UCAAGGGGCGGGGACAGGUCAAACUCGGCCUGUAACUGGAUGCAAUCACACACCCAGGGCUCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUU .(((((((((((((((((((......)))))))..((((.((........)).))))..)))).............(((.(((((...(((......)))..)))))))))))))))).. ( -39.00) >DroSim_CAF1 28584 120 - 1 UCAAGGGGCGGGGACAGGUCAAACUCGGCCUGUAACUGGAUGCAAUCACACACCCAGGGCUCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUU .(((((((((((((((((((......)))))))..((((.((........)).))))..)))).............(((.(((((...(((......)))..)))))))))))))))).. ( -39.00) >DroEre_CAF1 35713 119 - 1 -CGAGGGGCGGAGACAGGUCAAACUCGGCCUGUAACUGGAUGCAAUCACACACCCAGGGCUCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUU -(((((((((((((((((((......)))))))..((((.((........)).))))..)))).............(((.(((((...(((......)))..)))))))))))))))).. ( -39.50) >DroYak_CAF1 1768 120 - 1 UCAAGGGGCGGGGACCGUUCAAACUCGGCCUGUAACUGGAUGCAAUCACACACCCUGGGUCCUAAUACACAGAAACGCCAUAAACAAAACGUCCAAUUGUCAGUUUAGGCGCCUUUUGUU .((((((((.(((.(((........))))))(((...((((.((...........)).))))...)))........(((.(((((...(((......)))..)))))))))))))))).. ( -33.70) >consensus UCAAGGGGCGGGGACAGGUCAAACUCGGCCUGUAACUGGAUGCAAUCACACACCCAGGGCUCUAAUACACAGACACGCCAUAAACAAAGCGUCCAAUUGUCAGUUUAGGCGCCUUUUGUU .(((((((((((((((((((......)))))))..((((.((........)).))))..)))).............(((.(((((...(((......)))..)))))))))))))))).. (-35.62 = -35.54 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:38 2006