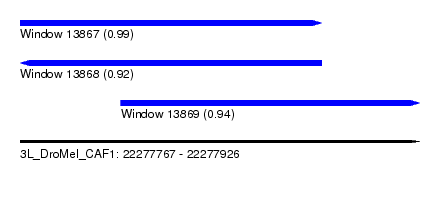
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,277,767 – 22,277,926 |
| Length | 159 |
| Max. P | 0.991027 |

| Location | 22,277,767 – 22,277,887 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.42 |
| Mean single sequence MFE | -42.86 |
| Consensus MFE | -39.86 |
| Energy contribution | -40.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.991027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22277767 120 + 23771897 ACGCGCUGCCGAGUGAAAUAUGUUAAUUAAAUUAAGGGCUGGGUGGUGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCA .......((((((((.................((((((((((((.((..........)).)).)))))).))))(((((((((..(..........)..))))))))).)))..))))). ( -43.60) >DroSec_CAF1 36149 120 + 1 ACGCGCUGCCGAGUGAAAUAUGUUAAUUAAAUUAAGGGCUGGUUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCA .......((((((((....................((((.((((((....(((.....)))..)))))))))).(((((((((..(..........)..))))))))).)))..))))). ( -41.70) >DroSim_CAF1 27750 120 + 1 ACGCGCUGCCGAGUGAAAUAUGUUAAUUAAAUUAAGGGCUGGGUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCA .......((((((((.................((((((((((((.(............).)).)))))).))))(((((((((..(..........)..))))))))).)))..))))). ( -41.50) >DroEre_CAF1 34917 120 + 1 ACCCGCUGCCGAUUGAAAUAUGUUAAUUAAAUUAAGGGCUGGGUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCA .(((((((((.((((.................((((((((((((.(............).)).)))))).))))(((....))).)))).)))))))))....((((.........)))) ( -46.00) >DroYak_CAF1 964 120 + 1 ACUCGCUGCCGAGUGAAAUAUGUUAAUUAAAUUAAGGGCUGGGUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCA .......((((((((.................((((((((((((.(............).)).)))))).))))(((((((((..(..........)..))))))))).)))..))))). ( -41.50) >consensus ACGCGCUGCCGAGUGAAAUAUGUUAAUUAAAUUAAGGGCUGGGUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCA .......((((((((.................((((((((((.((.....(((.....))))))))))).))))(((((((((..(..........)..))))))))).)))..))))). (-39.86 = -40.06 + 0.20)



| Location | 22,277,767 – 22,277,887 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.42 |
| Mean single sequence MFE | -36.86 |
| Consensus MFE | -35.34 |
| Energy contribution | -35.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22277767 120 - 23771897 UGCCGAAGGUGUUGGCAUGCCCCCGCUGCCCAUUGUUGGCAUGCCACAAGCAGCUGGUGUCGCUUUUCGAUUCACCACCCAGCCCUUAAUUUAAUUAACAUAUUUCACUCGGCAGCGCGU ((((((((.((((((((((((...((........)).)))))))))...))).))(((((((.....)))..))))................................))))))...... ( -35.70) >DroSec_CAF1 36149 120 - 1 UGCCGAAGGUGUUGGCAUGCCCCCGCUGCCCAUUGUUGGCAUGCCACAAGCAGCUGGUGUCGCUUUUCGAUUCCCCAACCAGCCCUUAAUUUAAUUAACAUAUUUCACUCGGCAGCGCGU ((((((..(((.(((((((((...((........)).))))))))).(((..((((((((((.....))))......)))))).)))..................)))))))))...... ( -37.60) >DroSim_CAF1 27750 120 - 1 UGCCGAAGGUGUUGGCAUGCCCCCGCUGCCCAUUGUUGGCAUGCCACAAGCAGCUGGUGUCGCUUUUCGAUUCCCCACCCAGCCCUUAAUUUAAUUAACAUAUUUCACUCGGCAGCGCGU ((((((..(((.(((((((((...((........)).))))))))).(((..(((((.((((.....)))).......))))).)))..................)))))))))...... ( -35.50) >DroEre_CAF1 34917 120 - 1 UGCCGAAGGUGUUGGCAUGCCCCCGCUGCCCAUUGUUGGCAUGCCACAAGCAGCUGGUGUCGCUUUUCGAUUCCCCACCCAGCCCUUAAUUUAAUUAACAUAUUUCAAUCGGCAGCGGGU ((((((.....))))))....(((((((((.((((.(((....))).(((..(((((.((((.....)))).......))))).)))..................)))).))))))))). ( -40.00) >DroYak_CAF1 964 120 - 1 UGCCGAAGGUGUUGGCAUGCCCCCGCUGCCCAUUGUUGGCAUGCCACAAGCAGCUGGUGUCGCUUUUCGAUUCCCCACCCAGCCCUUAAUUUAAUUAACAUAUUUCACUCGGCAGCGAGU ((((((..(((.(((((((((...((........)).))))))))).(((..(((((.((((.....)))).......))))).)))..................)))))))))...... ( -35.50) >consensus UGCCGAAGGUGUUGGCAUGCCCCCGCUGCCCAUUGUUGGCAUGCCACAAGCAGCUGGUGUCGCUUUUCGAUUCCCCACCCAGCCCUUAAUUUAAUUAACAUAUUUCACUCGGCAGCGCGU ((((((..((..(((((((((...((........)).)))))))))...)).(((((.((((.....)))).......))))).........................))))))...... (-35.34 = -35.34 + -0.00)


| Location | 22,277,807 – 22,277,926 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -43.44 |
| Consensus MFE | -42.06 |
| Energy contribution | -41.42 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22277807 119 + 23771897 GGGUGGUGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCAGCGGCAAAUUCUUCCUCUACCCCCCUU-CUUCGCACUUGA ((((((.((((((.....)))..((.(((((((((((((((((..(..........)..)))))))).))).....)))))))).......)))..)))))).....-............ ( -40.00) >DroSec_CAF1 36189 120 + 1 GGUUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCAGCGGCAAAUUCCUCCUCCAACCCCCUCCCUUCGCACUUGA (((((((((.(((.....)))..((.(((((((((((((((((..(..........)..)))))))).))).....))))))))........))).)))))).................. ( -43.20) >DroSim_CAF1 27790 120 + 1 GGGUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCAGCGGCAAAUUCCUCCUCCACCCCCCUCCCUGCGCACUUGA (((((((((.(((.....)))..((.(((((((((((((((((..(..........)..)))))))).))).....))))))))........))).)))))).................. ( -45.60) >DroEre_CAF1 34957 115 + 1 GGGUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCAGC----AAUGCUUCCUCCACCCCU-GCCCUUCUCCCUUGA (((((((((((((.....))).....(((((((((((((((((..(..........)..)))))))).))).....))))))----.....)))).))))))..-............... ( -45.40) >DroYak_CAF1 1004 114 + 1 GGGUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCAGC----AAUUCUUCCCCUACCUCU--CCCUUCUCACUUGA (((((((((((((.....))).....(((((((((((((((((..(..........)..)))))))).))).....))))))----.....)))))).))))..--.............. ( -43.00) >consensus GGGUGGGGAAUCGAAAAGCGACACCAGCUGCUUGUGGCAUGCCAACAAUGGGCAGCGGGGGCAUGCCAACACCUUCGGCAGCGGCAAAUUCUUCCUCCACCCCCCUCCCUUCGCACUUGA ((((((((..(((.....))).....(((((((((((((((((..(..........)..)))))))).))).....))))))............)))))))).................. (-42.06 = -41.42 + -0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:30 2006