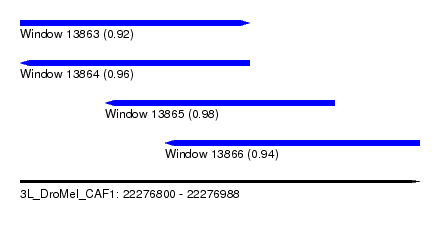
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,276,800 – 22,276,988 |
| Length | 188 |
| Max. P | 0.984212 |

| Location | 22,276,800 – 22,276,908 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.22 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22276800 108 + 23771897 CUACUGUCGCCUGUUUGCCUGCCAGUUGGAGUCCGCAUCCACAUCCAC------AUCCGCAUGCACAUCCGC------ACUCCCAUCCUCGUCCGCAUUUAUAUCCACAUGCAGACAGGC ........((((((((((.((......(((((.((.((...(((.(..------....).)))...)).)).------))))).......(....)...........)).)))))))))) ( -24.30) >DroSec_CAF1 35184 96 + 1 CUGCUGUCGCCUGUUUGCCUGCCAGUUGGCAUCCGCAUCCACAUCCAC------AUC------------CGC------AUCCACAUCCCCGUCCGCAUUUAUAUCCACAUGCAGAUAGGC ........((((((((((.((((....)))).................------...------------.((------....((......))..))..............)))))))))) ( -20.30) >DroSim_CAF1 26776 108 + 1 CUGCUGUCGCCUGUUUGCCUGCCAGUUGGCAUCCGCAUCCACAUCCAC------AUCCGCAUGCACAUCCGC------AUCCACAUCCCCGUCCGCAUUUAUAUCCACAUGCAGACAGGC ........((((((((((.((((....))))...((............------....(.((((......))------)).)((......))..))..............)))))))))) ( -26.80) >DroEre_CAF1 33978 120 + 1 CUGCUGUCGCCUGUUUGCCUGCCAGUUGGCAUCCGCAUCCACAUCCACAUCCACAUCCGCAUGCAGAUCCGCAUGCACAUCCGCAUCCCCAUCCGCAUUUAUAUCCUCAUGCAGACAGGC ........((((((((((.((((....))))...((......................((((((......))))))......(......)....))..............)))))))))) ( -32.50) >DroYak_CAF1 33 96 + 1 CUGCUGUCGCCUGUUUGCCUGCCAGUUGGCAUCCGCAUCCACAUCC------------------------UUAUGCACAUCCGCAUCCCCAUCCGCAUUUCUAUCCACAUGCAGACAGGC ........((((((((((.((((....))))...............------------------------..((((......))))........................)))))))))) ( -24.70) >consensus CUGCUGUCGCCUGUUUGCCUGCCAGUUGGCAUCCGCAUCCACAUCCAC______AUCCGCAUGCA_AUCCGC______AUCCACAUCCCCGUCCGCAUUUAUAUCCACAUGCAGACAGGC ........((((((((((.((((....))))...((..........................................................))..............)))))))))) (-19.34 = -19.22 + -0.12)

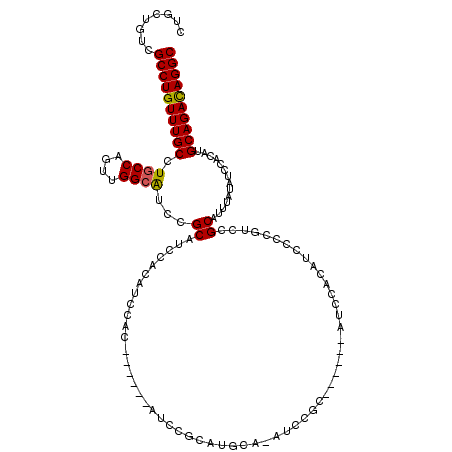

| Location | 22,276,800 – 22,276,908 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.86 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22276800 108 - 23771897 GCCUGUCUGCAUGUGGAUAUAAAUGCGGACGAGGAUGGGAGU------GCGGAUGUGCAUGCGGAU------GUGGAUGUGGAUGCGGACUCCAACUGGCAGGCAAACAGGCGACAGUAG .(((((((((((..........)))))))).)))...(((((------.((.((.(((((.((...------.)).))))).)).)).))))).((((...(.(.....).)..)))).. ( -39.50) >DroSec_CAF1 35184 96 - 1 GCCUAUCUGCAUGUGGAUAUAAAUGCGGACGGGGAUGUGGAU------GCG------------GAU------GUGGAUGUGGAUGCGGAUGCCAACUGGCAGGCAAACAGGCGACAGCAG .((..(((((((..........)))))))..))........(------((.------------..(------((.(.(.((..(((...((((....)))).)))..)).)).)))))). ( -30.10) >DroSim_CAF1 26776 108 - 1 GCCUGUCUGCAUGUGGAUAUAAAUGCGGACGGGGAUGUGGAU------GCGGAUGUGCAUGCGGAU------GUGGAUGUGGAUGCGGAUGCCAACUGGCAGGCAAACAGGCGACAGCAG .(((((((((((..........)))))))))))..((((.((------(((....))))).)...(------((.(.(.((..(((...((((....)))).)))..)).)).)))))). ( -39.60) >DroEre_CAF1 33978 120 - 1 GCCUGUCUGCAUGAGGAUAUAAAUGCGGAUGGGGAUGCGGAUGUGCAUGCGGAUCUGCAUGCGGAUGUGGAUGUGGAUGUGGAUGCGGAUGCCAACUGGCAGGCAAACAGGCGACAGCAG .(((((((((((..........)))))))))))..((((.((.((((((((....)))))))).)).)...(((.(.(.((..(((...((((....)))).)))..)).)).)))))). ( -46.80) >DroYak_CAF1 33 96 - 1 GCCUGUCUGCAUGUGGAUAGAAAUGCGGAUGGGGAUGCGGAUGUGCAUAA------------------------GGAUGUGGAUGCGGAUGCCAACUGGCAGGCAAACAGGCGACAGCAG ((((((.(((.(((...((....))(((...((.((.((.((.(((((..------------------------..))))).)).)).)).))..)))))).))).))))))........ ( -32.40) >consensus GCCUGUCUGCAUGUGGAUAUAAAUGCGGACGGGGAUGCGGAU______GCGGAU_UGCAUGCGGAU______GUGGAUGUGGAUGCGGAUGCCAACUGGCAGGCAAACAGGCGACAGCAG .(((((((((((..........))))))))))).......................................((.(.(.((..(((...((((....)))).)))..)).)).))..... (-25.82 = -26.86 + 1.04)


| Location | 22,276,840 – 22,276,948 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.66 |
| Mean single sequence MFE | -43.74 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.76 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22276840 108 - 23771897 GGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUGUCUGCAUGUGGAUAUAAAUGCGGACGAGGAUGGGAGU------GCGGAUGUGCAUGCGGAU------GUGGAUGU .(((((((((((.(((((((.(((..(((...........))).((((((((..........)))))))))))..)))))((------(((....))))))).)))------)))))))) ( -45.00) >DroSec_CAF1 35224 96 - 1 GGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUAUCUGCAUGUGGAUAUAAAUGCGGACGGGGAUGUGGAU------GCG------------GAU------GUGGAUGU .(((((((((..((((.(((.....))).))))..(((((((((.(((((((..........)))))))..))).)))))))------)))------------)))------))...... ( -37.70) >DroSim_CAF1 26816 108 - 1 GGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUCCGCCUGUCUGCAUGUGGAUAUAAAUGCGGACGGGGAUGUGGAU------GCGGAUGUGCAUGCGGAU------GUGGAUGU .(((((((((((.((...((.....))(((..(((.((((((((((((((((..........)))))))))))....)))).------).)))..)))..)).)))------)))))))) ( -45.20) >DroEre_CAF1 34018 120 - 1 GGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUGUCUGCAUGAGGAUAUAAAUGCGGAUGGGGAUGCGGAUGUGCAUGCGGAUCUGCAUGCGGAUGUGGAUGUGGAUGU .(((((((((((((((.(((.....))).))))........(((((((((((..........))))))))))))))))))))))((((.((.(((..(((....)))..))).)).)))) ( -52.40) >DroYak_CAF1 73 96 - 1 GGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUGUCUGCAUGUGGAUAGAAAUGCGGAUGGGGAUGCGGAUGUGCAUAA------------------------GGAUGU .(((((((((((((((.(((.....))).))))........(((((((((((..........))))))))))))))))))))))((((..------------------------..)))) ( -38.40) >consensus GGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUGUCUGCAUGUGGAUAUAAAUGCGGACGGGGAUGCGGAU______GCGGAU_UGCAUGCGGAU______GUGGAUGU ...(((((((((((((.(((.....))).))))........(((((((((((..........))))))))))))))))))))...................................... (-30.80 = -30.76 + -0.04)

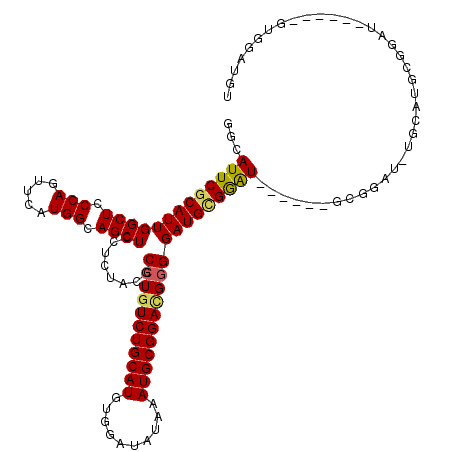
| Location | 22,276,868 – 22,276,988 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -43.28 |
| Consensus MFE | -41.36 |
| Energy contribution | -41.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22276868 120 - 23771897 UCCUGCUUCUUGCCUCCGACUGCGCUUUAAGUGGUCGCUUGGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUGUCUGCAUGUGGAUAUAAAUGCGGACGAGGAUGGGA (((((((((.((((..((((..(.......)..))))...))))((((((((((((.(((.....))).))))..((((((..((....))))))))....)))))))).)))).))))) ( -43.20) >DroSec_CAF1 35240 120 - 1 UCCUUCUUCUUGCCUCCGACUGCGCUUUAAGUGGUCGCUUGGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUAUCUGCAUGUGGAUAUAAAUGCGGACGGGGAUGUGG ..........((((..((((..(.......)..))))...))))..((((((((((.(((.....))).))))........((..(((((((..........)))))))..)))))))). ( -41.30) >DroSim_CAF1 26844 120 - 1 UCCUUCUUCUUGCCUCCGACUGCGCUUUAAGUGGUCGCUUGGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUCCGCCUGUCUGCAUGUGGAUAUAAAUGCGGACGGGGAUGUGG ..........((((..((((..(.......)..))))...))))..((((((((((.(((.....))).))))........(((((((((((..........))))))))))))))))). ( -46.10) >DroEre_CAF1 34058 120 - 1 UCCUGCUGCUUGCCACCGACCGAGCUUUAAGUGGUCGCUUGGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUGUCUGCAUGAGGAUAUAAAUGCGGAUGGGGAUGCGG ..........(((((.((((((.........))))))..)))))..((((((((((.(((.....))).))))........(((((((((((..........))))))))))))))))). ( -44.10) >DroYak_CAF1 89 120 - 1 UCCUCCUCCUUGCCUCCGACUGAGCUUUAAGUGGUCGCUUGGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUGUCUGCAUGUGGAUAGAAAUGCGGAUGGGGAUGCGG ..........((((..(((((..((.....)))))))...))))..((((((((((.(((.....))).))))........(((((((((((..........))))))))))))))))). ( -41.70) >consensus UCCUGCUUCUUGCCUCCGACUGCGCUUUAAGUGGUCGCUUGGCAUUCGCAUUGGCUCCCAGUUCAUGGCAGCUCCUCUACGCCUGUCUGCAUGUGGAUAUAAAUGCGGACGGGGAUGCGG ..........((((..(((((((.......)))))))...)))).(((((((((((.(((.....))).))))........(((((((((((..........)))))))))))))))))) (-41.36 = -41.72 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:27 2006