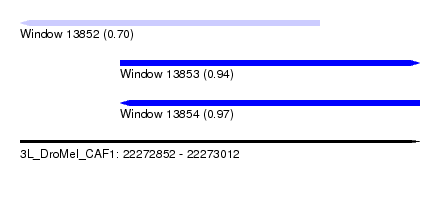
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,272,852 – 22,273,012 |
| Length | 160 |
| Max. P | 0.967048 |

| Location | 22,272,852 – 22,272,972 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -27.05 |
| Energy contribution | -27.05 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22272852 120 - 23771897 GAGCACUUUAAUGUGGCAGUGCGCUACAGUGGCACAUCGGUGCGGCAGAUGAAUUUUGCAUAUGACACUGCCUAAGUGUUCUUUCUGUCCGCCAAUUUGCAUUUUGCAUAUCGUUUAUAG ((((((((......((((((((((....)))((((....)))).(((((.....)))))......))))))).))))))))...((((..((.....(((.....)))....))..)))) ( -35.80) >DroSec_CAF1 31258 110 - 1 GAGCACUUUAAUGUGGCA----------GUGGCACAUCGGUGCGGCAGAUGAAUUUUGCAUAUGACACUGCCUAAGUGUUCUUUCUGUCCGCCAAUUUGCAUUUUGCAUAUCGUUUAUAG ((((((((......((((----------(((((((....)))).(((((.....)))))......))))))).))))))))...((((..((.....(((.....)))....))..)))) ( -35.60) >DroSim_CAF1 22834 110 - 1 GAGCACUUUAAUGUGGCA----------GUGGCACAUCGGUGCGGCAGAUGAAUUUUGCAUAUGACACUGCCUAAGUGUUCUUUCUGUCCGCCAAUUUGCAUUUUGCAUAUCGUUUAUAG ((((((((......((((----------(((((((....)))).(((((.....)))))......))))))).))))))))...((((..((.....(((.....)))....))..)))) ( -35.60) >DroEre_CAF1 30084 110 - 1 GGGCACUUUAAUGUGGCA----------GUGGCACAUUGGUGCGGCAGAUGGACUUUGCAUAUGACACUGCCCAAGUGUUCUUUCUGUCCGCCAAUUUGCAUUUUGAAUAUCGUUUAUAG .(.(((......))).).----------...(((.(((((((.((((((.(((...........(((((.....)))))))).))))))))))))).))).................... ( -32.80) >consensus GAGCACUUUAAUGUGGCA__________GUGGCACAUCGGUGCGGCAGAUGAAUUUUGCAUAUGACACUGCCUAAGUGUUCUUUCUGUCCGCCAAUUUGCAUUUUGCAUAUCGUUUAUAG ..(((.....(((((.((.((.....)).)).))))).((((.((((((.((((...(((........)))......))))..))))))))))....))).................... (-27.05 = -27.05 + 0.00)


| Location | 22,272,892 – 22,273,012 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -23.75 |
| Energy contribution | -23.56 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22272892 120 + 23771897 AACACUUAGGCAGUGUCAUAUGCAAAAUUCAUCUGCCGCACCGAUGUGCCACUGUAGCGCACUGCCACAUUAAAGUGCUCCGAAUGCCGGCCAAAACUUCACAUUAAAUCCAGGCCACAA ........((((...((....(((.........))).((((.((((((.((.((.....)).)).))))))...))))...)).))))((((....................)))).... ( -28.55) >DroSec_CAF1 31298 109 + 1 AACACUUAGGCAGUGUCAUAUGCAAAAUUCAUCUGCCGCACCGAUGUGCCAC----------UGCCACAUUAAAGUGCUCCGAAUGCCGGCCAAAACUUCACAUUAAAUCCAGGCCACA- ..(((((.(((((((......(((.........))).((((....)))))))----------))))......)))))...........((((....................))))...- ( -24.95) >DroSim_CAF1 22874 109 + 1 AACACUUAGGCAGUGUCAUAUGCAAAAUUCAUCUGCCGCACCGAUGUGCCAC----------UGCCACAUUAAAGUGCUCCGAAAGCCGGCCAAAACUUCACAUUAAAUCCAGGCCACA- ........(((((((......(((.........))).((((....)))))))----------))))........(.(((.....))))((((....................))))...- ( -25.55) >DroEre_CAF1 30124 109 + 1 AACACUUGGGCAGUGUCAUAUGCAAAGUCCAUCUGCCGCACCAAUGUGCCAC----------UGCCACAUUAAAGUGCCCCGAGUUCCGGCCAAAACUUCACAUUAAAUCCAGGCCACA- ...((((((((((((.(((((((..((.....))...)))....)))).)))----------)))(((......)))..))))))...((((....................))))...- ( -28.35) >consensus AACACUUAGGCAGUGUCAUAUGCAAAAUUCAUCUGCCGCACCGAUGUGCCAC__________UGCCACAUUAAAGUGCUCCGAAUGCCGGCCAAAACUUCACAUUAAAUCCAGGCCACA_ ........(((((((..............)).)))))((((.((((((.((...........)).))))))...))))..........((((....................)))).... (-23.75 = -23.56 + -0.19)



| Location | 22,272,892 – 22,273,012 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -33.97 |
| Energy contribution | -34.35 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.967048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22272892 120 - 23771897 UUGUGGCCUGGAUUUAAUGUGAAGUUUUGGCCGGCAUUCGGAGCACUUUAAUGUGGCAGUGCGCUACAGUGGCACAUCGGUGCGGCAGAUGAAUUUUGCAUAUGACACUGCCUAAGUGUU .(((((((..(((((......)))))..)))).))).....(((((((......((((((((((....)))((((....)))).(((((.....)))))......))))))).))))))) ( -40.40) >DroSec_CAF1 31298 109 - 1 -UGUGGCCUGGAUUUAAUGUGAAGUUUUGGCCGGCAUUCGGAGCACUUUAAUGUGGCA----------GUGGCACAUCGGUGCGGCAGAUGAAUUUUGCAUAUGACACUGCCUAAGUGUU -(((((((..(((((......)))))..)))).))).....(((((((......((((----------(((((((....)))).(((((.....)))))......))))))).))))))) ( -40.00) >DroSim_CAF1 22874 109 - 1 -UGUGGCCUGGAUUUAAUGUGAAGUUUUGGCCGGCUUUCGGAGCACUUUAAUGUGGCA----------GUGGCACAUCGGUGCGGCAGAUGAAUUUUGCAUAUGACACUGCCUAAGUGUU -..(((((..(((((......)))))..)))))........(((((((......((((----------(((((((....)))).(((((.....)))))......))))))).))))))) ( -39.60) >DroEre_CAF1 30124 109 - 1 -UGUGGCCUGGAUUUAAUGUGAAGUUUUGGCCGGAACUCGGGGCACUUUAAUGUGGCA----------GUGGCACAUUGGUGCGGCAGAUGGACUUUGCAUAUGACACUGCCCAAGUGUU -(.(((((..(((((......)))))..))))).)......((((((....((.((((----------(((((((....)))).(((((.....)))))......))))))))))))))) ( -39.00) >consensus _UGUGGCCUGGAUUUAAUGUGAAGUUUUGGCCGGCAUUCGGAGCACUUUAAUGUGGCA__________GUGGCACAUCGGUGCGGCAGAUGAAUUUUGCAUAUGACACUGCCUAAGUGUU ...(((((..(((((......)))))..))))).........(((((...(((((.((.((.....)).)).))))).))))).(((((.....)))))....((((((.....)))))) (-33.97 = -34.35 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:15 2006