
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,270,502 – 22,270,697 |
| Length | 195 |
| Max. P | 0.999890 |

| Location | 22,270,502 – 22,270,622 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -32.28 |
| Energy contribution | -33.65 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22270502 120 + 23771897 ACUUAAAUUCACGUACGAGGACCGAAAUGGCCACAGCUCUUACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCAAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUA ..................((...((..((((((.((.......)).))))))...))..(((....)))....)).(((((((((((...((((....))))..))).)))).))))... ( -34.00) >DroSec_CAF1 28920 120 + 1 ACUUAAAUUCACGUACGAGGACCGAAAUGGCCACAGCUCUUACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUA ........(((.((((..((.((.....))))(((((..((..(((((((((.......(((....)))......(((((....))))).)))))))))..))..)).)))))))))).. ( -36.50) >DroSim_CAF1 20508 120 + 1 ACUUAAAUUCACGUACGAGGACCGAAAUGACCACAGCUCUUACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUA ........(((.((((((((...((..((....))..))...))).((((((..((((.(((....)))......(((((....))))).))))..))))))........)))))))).. ( -33.40) >DroEre_CAF1 27811 119 + 1 ACUUAAAUUCACGUACCAGGACCGGAAUGGCCACAGCUC-UACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACGGCUAUAUGGCCGUGACCAAAUGCAUGUGUGCUGAUA ........(((.((((((((.(..((.((((((.((...-...)).))))))...))..).))...((((..((((((((....))))..))))..)).)).......)).))))))).. ( -37.00) >consensus ACUUAAAUUCACGUACGAGGACCGAAAUGGCCACAGCUCUUACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUA ........(((.((((..((.((.....))))(((((......(((((((((.......(((....)))......(((((....))))).)))))))))......)).)))))))))).. (-32.28 = -33.65 + 1.37)


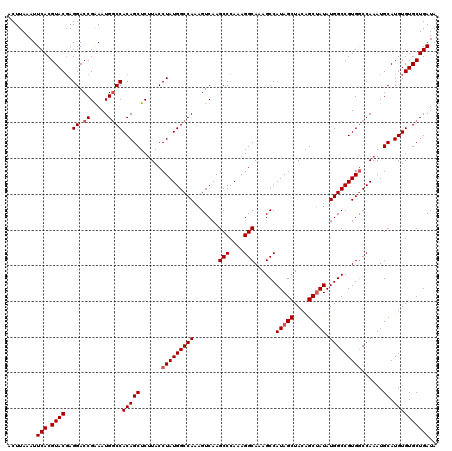
| Location | 22,270,502 – 22,270,622 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -40.76 |
| Energy contribution | -40.20 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22270502 120 - 23771897 UAUCAGCACACAUGCAUUUGGCCACGGCCAUAUUGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUAAGAGCUGUGGCCAUUUCGGUCCUCGUACGUGAAUUUAAGU ........(((.(((....(((...(((((((..((....)))))))))..)))...(((((.((...(((((((((.......)))))))))..)))))))..))).)))......... ( -41.90) >DroSec_CAF1 28920 120 - 1 UAUCAGCACACAUGCAUUUGGCCACGGCCAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUAAGAGCUGUGGCCAUUUCGGUCCUCGUACGUGAAUUUAAGU ........(((.(((....(((...(((((..((((....)))))))))..)))...(((((.((...(((((((((.......)))))))))..)))))))..))).)))......... ( -42.40) >DroSim_CAF1 20508 120 - 1 UAUCAGCACACAUGCAUUUGGCCACGGCCAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUAAGAGCUGUGGUCAUUUCGGUCCUCGUACGUGAAUUUAAGU ........(((.(((....(((...(((((..((((....)))))))))..)))...(((((.((...(((((((((.......)))))))))..)))))))..))).)))......... ( -39.70) >DroEre_CAF1 27811 119 - 1 UAUCAGCACACAUGCAUUUGGUCACGGCCAUAUAGCCGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUA-GAGCUGUGGCCAUUCCGGUCCUGGUACGUGAAUUUAAGU .....(((....)))(((((.(((((((((....((((..(((((((((((((((...((((.......))))..)))))-)))))))))).....))))..)))).)))))...))))) ( -45.90) >consensus UAUCAGCACACAUGCAUUUGGCCACGGCCAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUAAGAGCUGUGGCCAUUUCGGUCCUCGUACGUGAAUUUAAGU ........(((.(((....(((...(((((((..((....)))))))))..)))...(((((.((...(((((((((.......))))))))).)).)))))..))).)))......... (-40.76 = -40.20 + -0.56)

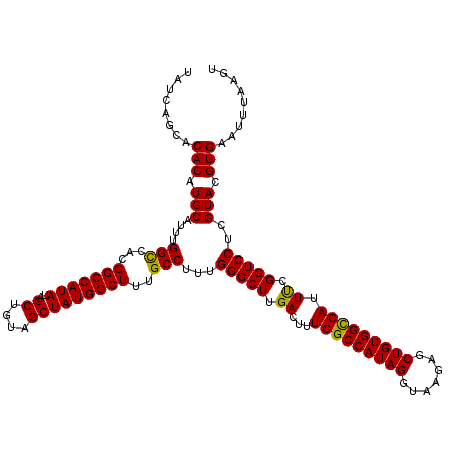
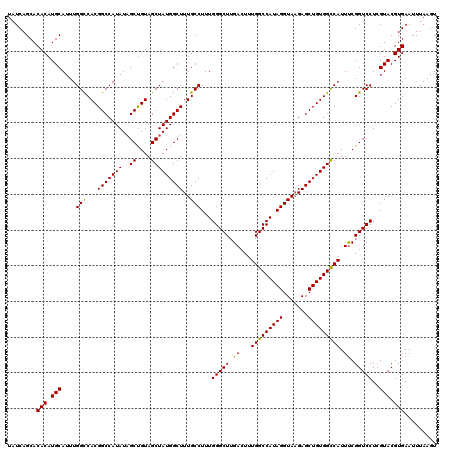
| Location | 22,270,542 – 22,270,662 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.04 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -31.43 |
| Energy contribution | -33.94 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22270542 120 + 23771897 UACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCAAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUAUAUGCACACUCUCUCGCUGGCCACACAUACGAUAUAUACG .......(((((.......(((....)))..(((....))).........)))))(((((((.......((((((........))))))........)))))))................ ( -38.56) >DroSec_CAF1 28960 119 + 1 UACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUAUAUGCCCACUCUCCCGCUGGCCACACAUACGAUAUAUAC- .......(((((.......(((....)))......(((((....))))).)))))(((((((.......(((.((........)).)))........)))))))...............- ( -36.86) >DroSim_CAF1 20548 119 + 1 UACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUAUAUGCACACUCUCCCGCUGGCCACACAUACGAUAUAUAC- .......(((((.......(((....)))......(((((....))))).)))))(((((((.......((((((........))))))........)))))))...............- ( -41.66) >DroEre_CAF1 27850 107 + 1 UACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACGGCUAUAUGGCCGUGACCAAAUGCAUGUGUGCUGAUAUAUGU------------UGUACGAGUAUAUG-UAUGUUCG ....((((((((.......(((....)))......(((((....))))).)))))))).....((((((((((((..........------------.))))....)))))-)))..... ( -31.10) >consensus UACCUAUGGCCAAAGUCAAGCCCAAAGGCAAAGCCAUAGCUACAGCUAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUAUAUGCACACUCUCCCGCUGGCCACACAUACGAUAUAUAC_ .......(((((.......(((....)))......(((((....))))).)))))(((((((.......((((((........))))))........)))))))................ (-31.43 = -33.94 + 2.50)

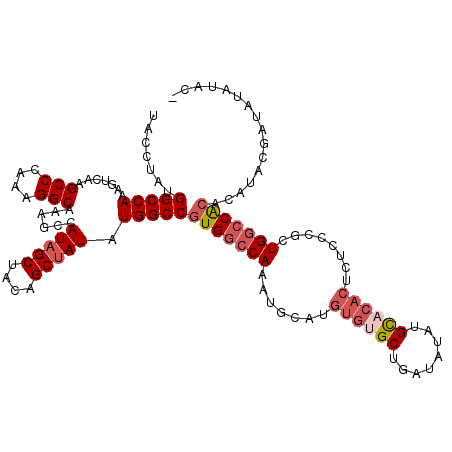

| Location | 22,270,542 – 22,270,662 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.04 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -31.68 |
| Energy contribution | -34.25 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22270542 120 - 23771897 CGUAUAUAUCGUAUGUGUGGCCAGCGAGAGAGUGUGCAUAUAUCAGCACACAUGCAUUUGGCCACGGCCAUAUUGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUA (((((.....))))).(((((((((....).((((((........))))))......))))))))(((((((..((....))))))))).(((((...((((.......))))..))))) ( -45.30) >DroSec_CAF1 28960 119 - 1 -GUAUAUAUCGUAUGUGUGGCCAGCGGGAGAGUGGGCAUAUAUCAGCACACAUGCAUUUGGCCACGGCCAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUA -.....((((((((((((((((.((......)).))).........)))))))))...((((((.(((((..((((....)))))))))..(((....))).......))))))..)))) ( -42.80) >DroSim_CAF1 20548 119 - 1 -GUAUAUAUCGUAUGUGUGGCCAGCGGGAGAGUGUGCAUAUAUCAGCACACAUGCAUUUGGCCACGGCCAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUA -...............((((((((..(.(..((((((........)))))).).)..))))))))(((((..((((....))))))))).(((((...((((.......))))..))))) ( -45.10) >DroEre_CAF1 27850 107 - 1 CGAACAUA-CAUAUACUCGUACA------------ACAUAUAUCAGCACACAUGCAUUUGGUCACGGCCAUAUAGCCGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUA ........-..............------------..................((...(((..(((((......)))))..)))..))..(((((...((((.......))))..))))) ( -25.50) >consensus _GUAUAUAUCGUAUGUGUGGCCAGCGGGAGAGUGUGCAUAUAUCAGCACACAUGCAUUUGGCCACGGCCAUAUAGCUGUAGCUAUGGCUUUGCCUUUGGGCUUGACUUUGGCCAUAGGUA ...............(((((((((.......((((((........))))))......)))))))))((((((..((....))))))))..(((((...((((.......))))..))))) (-31.68 = -34.25 + 2.56)



| Location | 22,270,582 – 22,270,697 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -36.69 |
| Consensus MFE | -23.44 |
| Energy contribution | -24.62 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.64 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22270582 115 + 23771897 UACAGCAAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUAUAUGCACACUCUCUCGCUGGCCACACAUACGAUAUAUACGA---UAUAUACACGUGCAUAUGUAU--GUAUAUGUCUGGC ............((((((((((.......((((((........))))))........)))))))......(((((((((.(---((((((........)))))))--))))))))).))) ( -40.46) >DroSec_CAF1 29000 105 + 1 UACAGCUAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUAUAUGCCCACUCUCCCGCUGGCCACACAUACGAUAUAUAC------------CC-UACAUAUGUAU--GUAUAUGUCUGGC ....((((..(((((((((......((((((((....))))))))........)))).))))).......(((((((((------------..-(((....))).--))))))))))))) ( -33.14) >DroSim_CAF1 20588 106 + 1 UACAGCUAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUAUAUGCACACUCUCCCGCUGGCCACACAUACGAUAUAUAC------------CCGUGCAUAUGUAU--GUAUAUGUCUGGC ............((((((((((.......((((((........))))))........)))))))......(((((((((------------..((((....))))--))))))))).))) ( -38.26) >DroEre_CAF1 27890 107 + 1 UACGGCUAUAUGGCCGUGACCAAAUGCAUGUGUGCUGAUAUAUGU------------UGUACGAGUAUAUG-UAUGUUCGAGUGUGUAUACACCUACGUAUGUAUACGAGUGUGUCUGGC (((((((....))))))).(((.((((((.(((((..........------------.))))).(((((((-(((((..(.((((....))))).))))))))))))..)))))).))). ( -34.90) >consensus UACAGCUAUAUGGCCGUGGCCAAAUGCAUGUGUGCUGAUAUAUGCACACUCUCCCGCUGGCCACACAUACGAUAUAUAC____________ACGUACAUAUGUAU__GUAUAUGUCUGGC ............((((((((((.......((((((........))))))........)))))))......(((((((((..............((((....))))..))))))))).))) (-23.44 = -24.62 + 1.19)

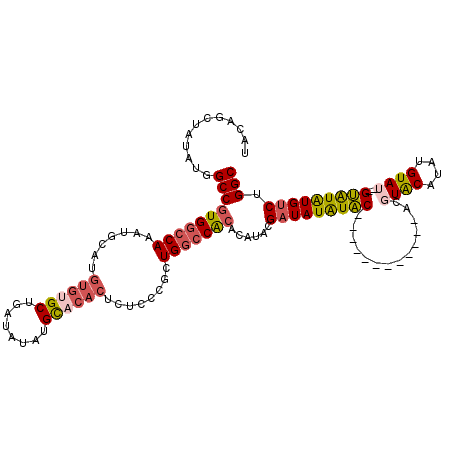

| Location | 22,270,582 – 22,270,697 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -18.91 |
| Energy contribution | -20.41 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22270582 115 - 23771897 GCCAGACAUAUAC--AUACAUAUGCACGUGUAUAUA---UCGUAUAUAUCGUAUGUGUGGCCAGCGAGAGAGUGUGCAUAUAUCAGCACACAUGCAUUUGGCCACGGCCAUAUUGCUGUA ((((.((((((..--((((((((((....)))))).---..)))).....)))))).))))((((((....((((((........)))))).......((((....))))..)))))).. ( -40.50) >DroSec_CAF1 29000 105 - 1 GCCAGACAUAUAC--AUACAUAUGUA-GG------------GUAUAUAUCGUAUGUGUGGCCAGCGGGAGAGUGGGCAUAUAUCAGCACACAUGCAUUUGGCCACGGCCAUAUAGCUGUA ((((((.((((((--.(((....)))-..------------)))))).))((((((((((((.((......)).))).........)))))))))...)))).(((((......))))). ( -36.20) >DroSim_CAF1 20588 106 - 1 GCCAGACAUAUAC--AUACAUAUGCACGG------------GUAUAUAUCGUAUGUGUGGCCAGCGGGAGAGUGUGCAUAUAUCAGCACACAUGCAUUUGGCCACGGCCAUAUAGCUGUA ((((.((((((..--....((((((....------------))))))...)))))).))))((((......((((((........)))))).......((((....))))....)))).. ( -37.60) >DroEre_CAF1 27890 107 - 1 GCCAGACACACUCGUAUACAUACGUAGGUGUAUACACACUCGAACAUA-CAUAUACUCGUACA------------ACAUAUAUCAGCACACAUGCAUUUGGUCACGGCCAUAUAGCCGUA ((((((.......((((((((......)))))))).............-..............------------..........(((....))).)))))).(((((......))))). ( -23.20) >consensus GCCAGACAUAUAC__AUACAUAUGCACGG____________GUAUAUAUCGUAUGUGUGGCCAGCGGGAGAGUGUGCAUAUAUCAGCACACAUGCAUUUGGCCACGGCCAUAUAGCUGUA ((((((..........(((((((((.........................)))))))))............((((((........)))))).....)))))).(((((......))))). (-18.91 = -20.41 + 1.50)

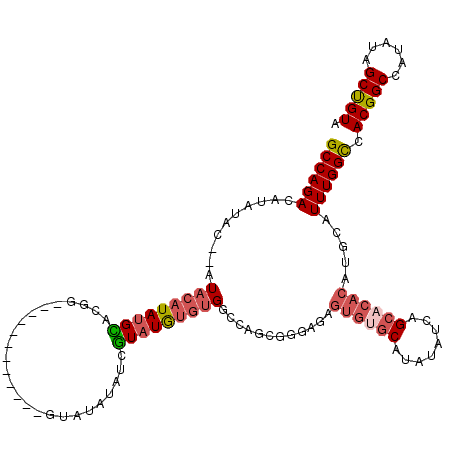

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:07 2006