
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,269,508 – 22,269,662 |
| Length | 154 |
| Max. P | 0.983763 |

| Location | 22,269,508 – 22,269,622 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -29.62 |
| Energy contribution | -30.07 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
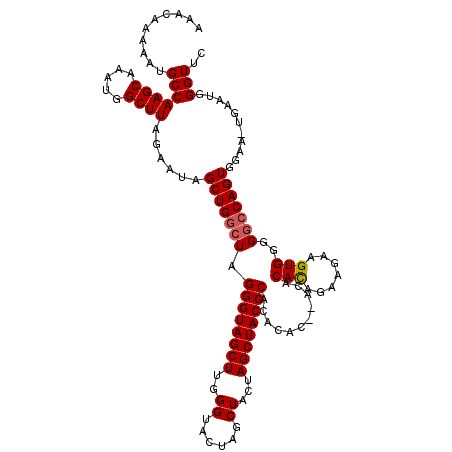
>3L_DroMel_CAF1 22269508 114 - 23771897 AAACAAAAAUGCCAAGCAAAUGGCUUAGAAUAGCUGGCUAGGGUAGCUUGGGUA---UCUACUAGCUACCCACACAU--ACACAUAGGAGAAGUGGGGGGCAGUAGAA-UGAAUGGGUUC .(((.......((((((...(((((.((.....))))))).....))))))...---((((((.((..(((((....--.(.....).....)))))..)))))))).-.......))). ( -33.50) >DroSec_CAF1 27901 120 - 1 AAACAAAAAUGCCAAGCAAAUGGCUUAGAAUAGCUGGUUAGGGUAGCUUGGGUACUAGCUCCUAGCUACCCACACACUCACACACAGAAGAAGUGGGGCCCAGUGGAAAGGAAUGGGUUC ..........(((((((.....))))......(((((...((((((((.((((....)).)).)))))))).....(((((...........)))))..)))))...........))).. ( -35.50) >DroSim_CAF1 19510 117 - 1 AAACAAAAAUGCCAAGCAAAUGGCUUAGAAUAGCUGGCUAGGGUAGCUUGGGUACUAGCUGCUAGCUACCCACACAC--ACACACAGAAGAAGUGGGGGCCAGUGGAA-UGAAUGGGUUC ..........(((((((.....))))......(((((((.((((((((..(((....)))...))))))))......--...(((.......)))..)))))))....-......))).. ( -35.20) >consensus AAACAAAAAUGCCAAGCAAAUGGCUUAGAAUAGCUGGCUAGGGUAGCUUGGGUACUAGCUACUAGCUACCCACACAC__ACACACAGAAGAAGUGGGGGCCAGUGGAA_UGAAUGGGUUC ..........(((((((.....))))......(((((((.((((((((..((......))...))))))))...........(((.......)))..)))))))...........))).. (-29.62 = -30.07 + 0.45)

| Location | 22,269,547 – 22,269,662 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -18.21 |
| Energy contribution | -19.84 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
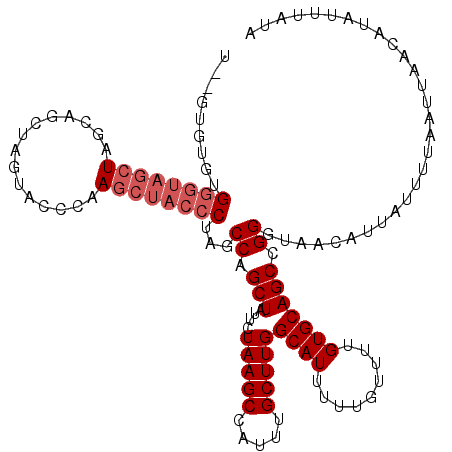
>3L_DroMel_CAF1 22269547 115 + 23771897 U--AUGUGUGGGUAGCUAGUAGA---UACCCAAGCUACCCUAGCCAGCUAUUCUAAGCCAUUUGCUUGGCAUUUUUGUUUUGUGCAGCCGGGUAGCAUUAUUUUAAUUAACGUAUUUAUA (--((((.((((((.(.....).---)))))).(((((((..((((((...............)).))))....((((.....))))..))))))).............)))))...... ( -32.46) >DroSec_CAF1 27941 120 + 1 UGAGUGUGUGGGUAGCUAGGAGCUAGUACCCAAGCUACCCUAACCAGCUAUUCUAAGCCAUUUGCUUGGCAUUUUUGUUUUGUGCAGCCGGGUAACAUUAUUUUAAUUAACAUACUUAUA (((((((((((((((((.((.(......))).))))))))...((.(((....(((((.....)))))((((.........))))))).))..................))))))))).. ( -37.70) >DroSim_CAF1 19549 118 + 1 U--GUGUGUGGGUAGCUAGCAGCUAGUACCCAAGCUACCCUAGCCAGCUAUUCUAAGCCAUUUGCUUGGCAUUUUUGUUUUGUGCAGCCGGGUAACAUUAUUUUAAUUAACAUAUUUAUA .--(((((((((((.((((...)))))))))..(.(((((..((((((...............)).))))....((((.....))))..))))).).............))))))..... ( -29.66) >DroEre_CAF1 26849 96 + 1 -------------------CCACUUGUACCCC-UCUACC----CCAGCUAUUCUAAGCCAUUGGCUUGGCAUUUUUGUUCUGUGCAGCCGGGUAACAUUAUUUUAAUUAACAUAUUUAUA -------------------.............-..((((----(..(((....((((((...))))))((((.........))))))).))))).......................... ( -18.10) >consensus U__GUGUGUGGGUAGCUAGCAGCUAGUACCCAAGCUACCCUAGCCAGCUAUUCUAAGCCAUUUGCUUGGCAUUUUUGUUUUGUGCAGCCGGGUAACAUUAUUUUAAUUAACAUAUUUAUA .........((((((((...............))))))))...((.(((....(((((.....)))))((((.........))))))).))............................. (-18.21 = -19.84 + 1.63)


| Location | 22,269,547 – 22,269,662 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -19.26 |
| Energy contribution | -20.95 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
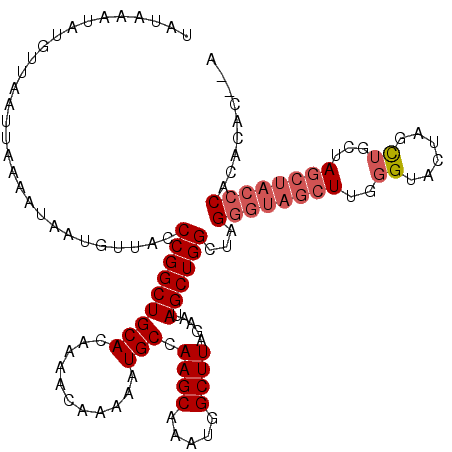
>3L_DroMel_CAF1 22269547 115 - 23771897 UAUAAAUACGUUAAUUAAAAUAAUGCUACCCGGCUGCACAAAACAAAAAUGCCAAGCAAAUGGCUUAGAAUAGCUGGCUAGGGUAGCUUGGGUA---UCUACUAGCUACCCACACAU--A ........................((((((((((................))).(((....((((......)))).))).))))))).((((((---.(.....).)))))).....--. ( -31.29) >DroSec_CAF1 27941 120 - 1 UAUAAGUAUGUUAAUUAAAAUAAUGUUACCCGGCUGCACAAAACAAAAAUGCCAAGCAAAUGGCUUAGAAUAGCUGGUUAGGGUAGCUUGGGUACUAGCUCCUAGCUACCCACACACUCA ....(((.(((..................(((((((..............((((......))))......)))))))...((((((((.((((....)).)).))))))))))).))).. ( -32.45) >DroSim_CAF1 19549 118 - 1 UAUAAAUAUGUUAAUUAAAAUAAUGUUACCCGGCUGCACAAAACAAAAAUGCCAAGCAAAUGGCUUAGAAUAGCUGGCUAGGGUAGCUUGGGUACUAGCUGCUAGCUACCCACACAC--A ........................((((((((((................))).(((....((((......)))).))).))))))).((((((((((...)))).)))))).....--. ( -32.09) >DroEre_CAF1 26849 96 - 1 UAUAAAUAUGUUAAUUAAAAUAAUGUUACCCGGCUGCACAGAACAAAAAUGCCAAGCCAAUGGCUUAGAAUAGCUGG----GGUAGA-GGGGUACAAGUGG------------------- .......................(((.((((..((((.(((.........((((......)))).........))).----.)))).-.))))))).....------------------- ( -23.17) >consensus UAUAAAUAUGUUAAUUAAAAUAAUGUUACCCGGCUGCACAAAACAAAAAUGCCAAGCAAAUGGCUUAGAAUAGCUGGCUAGGGUAGCUUGGGUACUAGCUGCUAGCUACCCACACAC__A .............................(((((((((...........))).((((.....)))).....))))))...((((((((..((......))...))))))))......... (-19.26 = -20.95 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:16:01 2006