
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,268,446 – 22,268,644 |
| Length | 198 |
| Max. P | 0.974677 |

| Location | 22,268,446 – 22,268,564 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -41.42 |
| Consensus MFE | -37.81 |
| Energy contribution | -37.75 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22268446 118 - 23771897 CACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGUAGCAGCGCUUGUUGCAACUUUAUUUACCUUUACGUAUGACAUAACUUGUGAAUAGAGAUACAAACGGGGCUUGCACGUAU--GUG ...(((((....))))).(((((((((((.((....))..))))))))))).((((((((((..(((.(.....).)))...)))))))))).((((.(((..(....).))).)--))) ( -39.00) >DroSec_CAF1 26748 118 - 1 CACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGCAGCAGCGCUUGUUGCAACUUUAUUUACCUUUACGUGGGACAUAACUUGUAAAUAGAGAUACAAACGGGGGUUGCACGUGU--GUG ((((((((....))))).((((((......))))))(((((.((((((....((((((((.(((......)))(((.....))))))))))).....)))))).)))))..))).--... ( -43.10) >DroSim_CAF1 17149 118 - 1 CACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGCAGCAGCGCUUGUUGCAACUUUAUUUACCUUUACGUGGGACAUAACUUGUAAAUAGAGAUACAAACGGGGGUUGCACGUGU--GUG ((((((((....))))).((((((......))))))(((((.((((((....((((((((.(((......)))(((.....))))))))))).....)))))).)))))..))).--... ( -43.10) >DroEre_CAF1 25701 120 - 1 CACGUCACUUCGGUGGCAUGCAACGAGCGAGUUACAGCAGCGUUUGUUGCAACUUUAUUUACCUUUAGGUGGGACAUAACUUGUAAAUAGAGAUACAAACGGGGGUUGCACGUGUGUGUG ...(((((....))))).(((((((((((.((....))..))))))))))).((((((((((....((((........))))))))))))))(((((.(((.........))).))))). ( -40.50) >consensus CACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGCAGCAGCGCUUGUUGCAACUUUAUUUACCUUUACGUGGGACAUAACUUGUAAAUAGAGAUACAAACGGGGGUUGCACGUGU__GUG ((((((((....))))).((((((......))))))(((((.((((((....((((((((((..(((.(.....).)))...)))))))))).....)))))).)))))........))) (-37.81 = -37.75 + -0.06)



| Location | 22,268,484 – 22,268,604 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -38.06 |
| Energy contribution | -38.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22268484 120 - 23771897 ACGGCAAACCACAAGCGCGCUUUGAGCAAAGGAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGUAGCAGCGCUUGUUGCAACUUUAUUUACCUUUACGUAUGACAUAAC ..((....)).((.(((.((.....))(((((..(((((....(((((....))))).(((((((((((.((....))..)))))))))))))))).....))))).))).))....... ( -38.70) >DroSec_CAF1 26786 120 - 1 ACGGCAAACCACAAGCGCGCUUUGAGCGAAGGAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGCAGCAGCGCUUGUUGCAACUUUAUUUACCUUUACGUGGGACAUAAC ........((((..(.(((((((...(....)..))))))).)(((((....))))).(((((((((((.((....))..))))))))))).................))))........ ( -43.30) >DroSim_CAF1 17187 120 - 1 ACGGCAAACCACAAGCGCGCUUUGAGCGAAGCAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGCAGCAGCGCUUGUUGCAACUUUAUUUACCUUUACGUGGGACAUAAC ...(((....(((((((((((((....)))))......(((..(((((....))))).((((((......))))))))))))))))))))...........(((......)))....... ( -45.00) >DroEre_CAF1 25741 120 - 1 ACGGCAAACCACAAGCGCGCUUUGAGCGGAGGAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUACAGCAGCGUUUGUUGCAACUUUAUUUACCUUUAGGUGGGACAUAAC ........((((...(((.......)))((((..(((((....(((((....))))).(((((((((((.((....))..)))))))))))))))).....))))...))))........ ( -39.90) >consensus ACGGCAAACCACAAGCGCGCUUUGAGCGAAGGAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGCAGCAGCGCUUGUUGCAACUUUAUUUACCUUUACGUGGGACAUAAC ........((((....((.......))(((((..(((((....(((((....))))).(((((((((((.((....))..)))))))))))))))).....)))))..))))........ (-38.06 = -38.25 + 0.19)

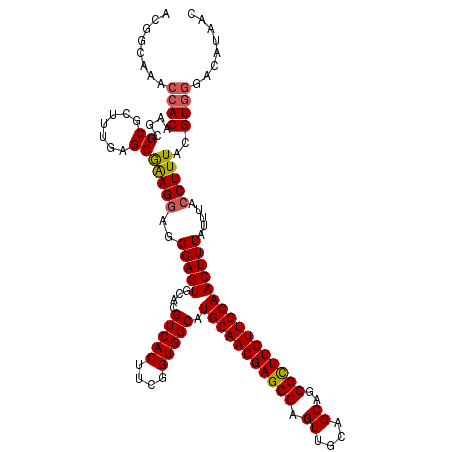

| Location | 22,268,524 – 22,268,644 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -35.96 |
| Energy contribution | -36.02 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22268524 120 - 23771897 GAAAAUUCAUUUCAAUUAGUGAAAUGGUUUUGGGAUUUGUACGGCAAACCACAAGCGCGCUUUGAGCAAAGGAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGUAGCAG ......((((((((.....))))))))..(((((.((((.....)))))).)))(((((((((...(....)..)))))))..(((((....))))).((((((......)))))))).. ( -36.10) >DroSec_CAF1 26826 120 - 1 GAAAAUUCAUUUCAAUUAGUGAAAUGGUUUUGGGAUUUGUACGGCAAACCACAAGCGCGCUUUGAGCGAAGGAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGCAGCAG ......((((((((.....))))))))..(((((.((((.....)))))).)))(((((((((...(....)..)))))))..(((((....))))).((((((......)))))))).. ( -40.80) >DroSim_CAF1 17227 120 - 1 GAAAAUUCAUUUCAAUUAGUGAAAUGGUUUUGGGAUUUGUACGGCAAACCACAAGCGCGCUUUGAGCGAAGCAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGCAGCAG ......((((((((.....))))))))..(((((.((((.....)))))).)))(((((((((..((...))..)))))))..(((((....))))).((((((......)))))))).. ( -40.50) >DroEre_CAF1 25781 120 - 1 GAAAAUUCAUUUCAAUUAGUGAAAUGGUUUUGGGAUUUGUACGGCAAACCACAAGCGCGCUUUGAGCGGAGGAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUACAGCAG ......((((((((.....))))))))..(((..((((((.((.....((.(...(.(((.....))))..).))...((((.(((((....))))).)))).)).))))))..)))... ( -36.00) >consensus GAAAAUUCAUUUCAAUUAGUGAAAUGGUUUUGGGAUUUGUACGGCAAACCACAAGCGCGCUUUGAGCGAAGGAGGGAGUGCACGUCACUUCGGUGGCAUGCAACGAGCGAGUUGCAGCAG ......((((((((.....))))))))..(((((.((((.....)))))).)))(((((((((...........)))))))..(((((....))))).((((((......)))))))).. (-35.96 = -36.02 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:59 2006