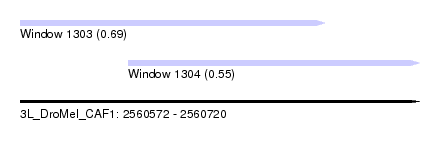
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,560,572 – 2,560,720 |
| Length | 148 |
| Max. P | 0.687844 |

| Location | 2,560,572 – 2,560,685 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.28 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -7.44 |
| Energy contribution | -7.88 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
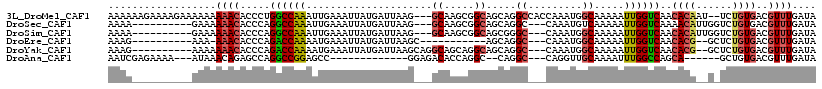
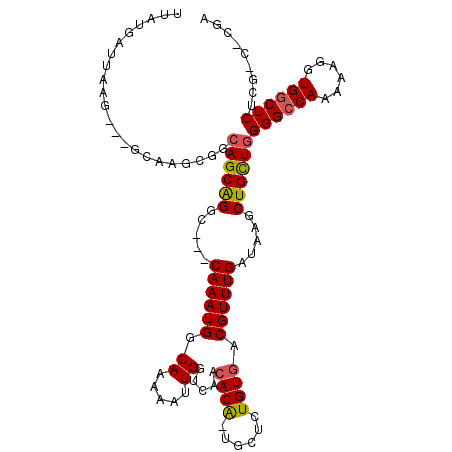
>3L_DroMel_CAF1 2560572 113 + 23771897 AAAAAAGAAAAGAAAAAAAAACACCCUGGCCAAAUUGAAAUUAUGAUUAAG---GCAAGCGGCAGCAGGCCACCAAAUGGCAAAAAUUGGUCAACACAAU--UCUGUGACGUUUGAUA ..................((((....(((((((.................(---((..((....))..))).((....))......))))))).((((..--..))))..)))).... ( -22.30) >DroSec_CAF1 22451 102 + 1 AAAA----------GAAAAAACACCCAGGCCAAAUUGAAAUUAUGAUUAAG---GCAAGCGGCAGCAGGC---CAAAUGUCAAAAAUUGGUCAAAACAUUGGUCUGUGACGUUUGAUA ....----------....((((..(((((((((.((((.....((((...(---((..((....))..))---)....))))........))))....)))))))).)..)))).... ( -23.12) >DroSim_CAF1 19428 102 + 1 AAAA----------GAAAAAACACCCAGGCCAAAUUGAAAUUAUGAUUAAG---GCAAGCGGCAGCGGGC---CAAAUGGCAAAAAUUGGUCAACACAUUGGUCUGUGACGUUUGAUA ....----------....((((..(((((((((..((.............(---((..((....))..))---)...((((........)))).))..)))))))).)..)))).... ( -25.20) >DroEre_CAF1 36687 91 + 1 AAAG----------AAA-AAACACCCAGACCAAAAUGAAAUUAUGAUUAAGC-----------AGCAGGC---CAAAUGGCAAAAAUUGGUCAACACG--GCUCUGUGACGUUUGAUA ....----------...-((((.....((((((..((...(((....))).)-----------)....((---(....))).....))))))..((((--....))))..)))).... ( -17.30) >DroYak_CAF1 21093 103 + 1 AAAG----------AAAAAAACACCCAGACCAAAAUGAAAUUAUGAUUAAGCAGGCAGCAGGCAGCAGGC---CAAAUGGCAAAAAUUGGUCAACACG--GCUCUGUGACGUUUGAUA ....----------....((((.....((((((.................((..((.....)).))..((---(....))).....))))))..((((--....))))..)))).... ( -21.00) >DroAna_CAF1 20513 91 + 1 AAUCGAGAAAA---AUAAACAGAGCCAGGCCGGAGCC-------------GGAGACACCAGGC--CAGGC---CAGGUUGCAAAAUUUGGCCAGCA------GCUGUGACGUUUGAUA .((((((....---....((((.((..((((((....-------------(....).)).)))--).(((---((((((....))))))))).)).------.))))....)))))). ( -31.00) >consensus AAAA__________AAAAAAACACCCAGGCCAAAUUGAAAUUAUGAUUAAG___GCAAGCGGCAGCAGGC___CAAAUGGCAAAAAUUGGUCAACACA__G_UCUGUGACGUUUGAUA ..................((((.....((((((.....................((.....)).....((.........)).....))))))..((((......))))..)))).... ( -7.44 = -7.88 + 0.45)
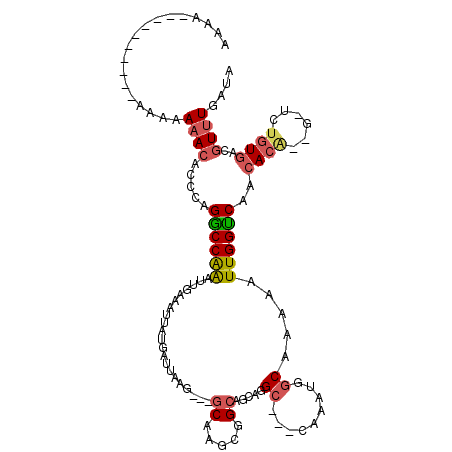
| Location | 2,560,612 – 2,560,720 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.47 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.26 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2560612 108 + 23771897 UUAUGAUUAAG---GCAAGCGGCAGCAGGCCACCAAAUGGCAAAAAUUGGUCAACACAAU--UCUGUGACGUUUGAUAAGCUGUUGGGGCCAAAAAGGUGACUCUCGCCCCGA ...........---.((((((...((((((((.....))))...(((((.......))))--)))))..))))))........(((((((.....((....))...))))))) ( -32.50) >DroSec_CAF1 22481 100 + 1 UUAUGAUUAAG---GCAAGCGGCAGCAGGC---CAAAUGUCAAAAAUUGGUCAAAACAUUGGUCUGUGACGUUUGAUAAGCUGCUGGGGCCAAAAAGGUGGCUCUC------- ...........---.....(((((((..(.---(((((((((......(..(((....)))..)..))))))))).)..)))))))((((((......))))))..------- ( -34.40) >DroSim_CAF1 19458 100 + 1 UUAUGAUUAAG---GCAAGCGGCAGCGGGC---CAAAUGGCAAAAAUUGGUCAACACAUUGGUCUGUGACGUUUGAUAAGCUGCUGGGGCCAAAAAGGUGGCUCUC------- ...........---.....(((((((..((---(....)))........(((((((((......))))....)))))..)))))))((((((......))))))..------- ( -31.00) >DroEre_CAF1 36716 97 + 1 UUAUGAUUAAGC-----------AGCAGGC---CAAAUGGCAAAAAUUGGUCAACACG--GCUCUGUGACGUUUGAUAAGCUGCUGGGGCCAAAAAGGUGGCUCUCGUCUCGA ....(((..(((-----------(((..((---(....)))........(((((.(((--.........))))))))..))))))(((((((......))))))).))).... ( -30.10) >DroYak_CAF1 21123 108 + 1 UUAUGAUUAAGCAGGCAGCAGGCAGCAGGC---CAAAUGGCAAAAAUUGGUCAACACG--GCUCUGUGACGUUUGAUAAGCUGCUGGGGCCAAAAAGGUGGCUCUCGCCUCGA ..........(.((((....((((((..((---(....)))........(((((.(((--.........))))))))..))))))(((((((......))))))).))))).. ( -36.50) >consensus UUAUGAUUAAG___GCAAGCGGCAGCAGGC___CAAAUGGCAAAAAUUGGUCAACACA_UGCUCUGUGACGUUUGAUAAGCUGCUGGGGCCAAAAAGGUGGCUCUCG_C_CGA ......................((((((.....((((((.((.....)).....((((......)))).)))))).....))))))((((((......))))))......... (-20.22 = -20.26 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:39 2006