
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,264,622 – 22,264,850 |
| Length | 228 |
| Max. P | 0.958993 |

| Location | 22,264,622 – 22,264,736 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -27.95 |
| Energy contribution | -29.07 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22264622 114 + 23771897 UUGUUGGUUUGCGUUUUAUUUAAAUUUGCUUCUUUCGGCUGCCUUUUUGCUC------GUGCGGUGGUUGUUUGUCGCCCGAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUUCGUG .....(((((((((((.....)))).(((((..(((((((((..((....((------(.((((..(....)..)))).)))......))..)))))))))))))).)).)))))..... ( -30.90) >DroSec_CAF1 22959 111 + 1 UUGUUGGUUUGCGUUUUAUUUAAAUUUGCU---UUCGGCUGCCUUUUUGCUC------GCGCGGUGGUUGUUUGUCGCCCGAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUGGCUG ..((..((((((((((.....)))).((((---(((((((((..((....((------(.((((..(....)..)))).)))......))..))))))).)))))).)).))))..)).. ( -36.20) >DroSim_CAF1 13176 114 + 1 UUGUUGGUUUGCGUUUUAUUUAAAUUUGCUUCUUUCGGCUGCCUUUUUGCUC------GCGCGGUGGUUGUUUGUCGCCCGAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUGGCUG ..((..((((((((((.....)))).(((((..(((((((((..((....((------(.((((..(....)..)))).)))......))..)))))))))))))).)).))))..)).. ( -36.20) >DroEre_CAF1 22016 120 + 1 UUGUUGGUUUGCGUUUUAUUUAAAUUUGCUUCUUUCGGCUGGCUUUUUGCUCCUGCUUGCUCGGUGGUUGUCUGUCGCCCGAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUGGCUG ..(..((...(((..((.....))..)))..))..)(((..((((...((...(((((..(((((.((((((..((....))......)))))).))))).))))).)).))))..))). ( -37.00) >consensus UUGUUGGUUUGCGUUUUAUUUAAAUUUGCUUCUUUCGGCUGCCUUUUUGCUC______GCGCGGUGGUUGUUUGUCGCCCGAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUGGCUG ..((((((((((((((.....)))).(((((..(((((((((......((........))((((..(....)..))))..............)))))))))))))).)).)))))))).. (-27.95 = -29.07 + 1.12)


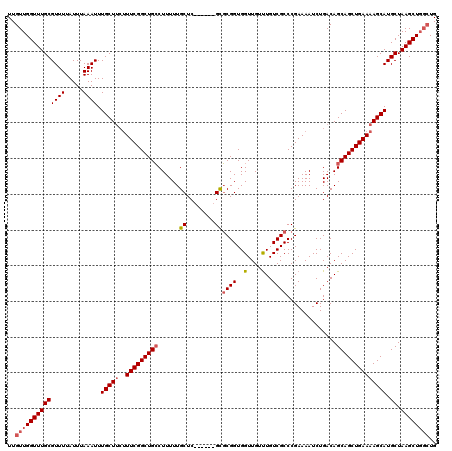
| Location | 22,264,662 – 22,264,776 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.02 |
| Mean single sequence MFE | -31.61 |
| Consensus MFE | -25.73 |
| Energy contribution | -26.04 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22264662 114 - 23771897 GCAAAUCAUUAAUUUGCUUUGAGCCAGAGCAAAAAGAAAGCACGAAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUCGGGCGACAAACAACCACCGCAC------GAGCAAAAAGGC (((((((.....(((((((((...)))))))))..((((((.....))))((((.(((....))))))).)).)))))(((.(((............))).)------))))........ ( -28.00) >DroSec_CAF1 22996 114 - 1 GCAAAUCAUUAAUUUGCUUUGAGCCAAAGCAAAAAGAAAGCAGCCAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUCGGGCGACAAACAACCACCGCGC------GAGCAAAAAGGC (((((((.....(((((((((...)))))))))..((.((((((.(((.......))).....)))))).)).)))))(((.(((............))).)------))))........ ( -33.30) >DroSim_CAF1 13216 114 - 1 GCAAAUCAUUAAUUUGCUUUGAGCCAAAGCAAAAAGAAAGCAGCCAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUCGGGCGACAAACAACCACCGCGC------GAGCAAAAAGGC (((((((.....(((((((((...)))))))))..((.((((((.(((.......))).....)))))).)).)))))(((.(((............))).)------))))........ ( -33.30) >DroEre_CAF1 22056 120 - 1 GCAAAUCAUUAAUUUGCUUUGAGCCAAAGCAAAAAGAAAGCAGCCAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUCGGGCGACAGACAACCACCGAGCAAGCAGGAGCAAAAAGCC ((..........(((((((((...)))))))))....((((.....)))).))..((((((..(((.((((..(....((((..............)))).)..)))).))).)))))). ( -31.84) >consensus GCAAAUCAUUAAUUUGCUUUGAGCCAAAGCAAAAAGAAAGCAGCCAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUCGGGCGACAAACAACCACCGCGC______GAGCAAAAAGGC (((((((.....(((((((((...)))))))))..((.((((((.(((.......))).....)))))).)).))))).(((..............)))...........))........ (-25.73 = -26.04 + 0.31)

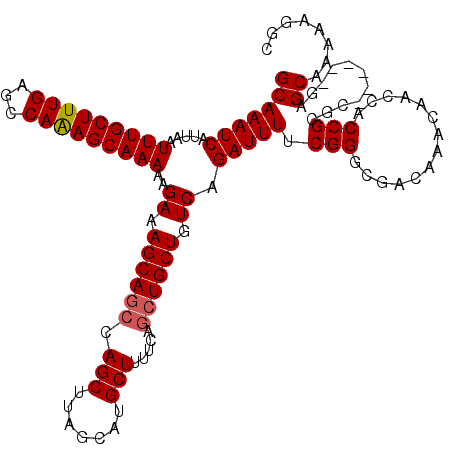
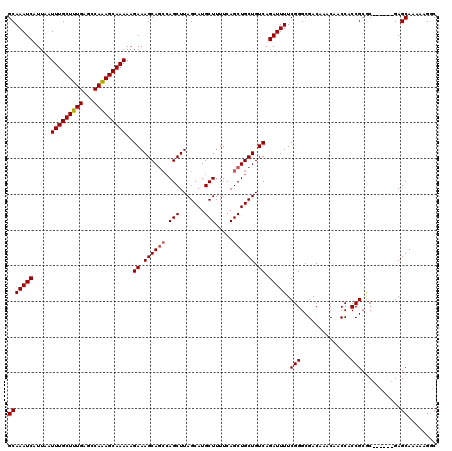
| Location | 22,264,696 – 22,264,816 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -28.83 |
| Energy contribution | -30.57 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22264696 120 + 23771897 GAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUUCGUGCUUUCUUUUUGCUCUGGCUCAAAGCAAAUUAAUGAUUUGCCAUCAAGCAAAUAAAUCAAAAGCUUCAAAGCAGAUCUCUG ....(((((.((((((((....))).))))((((((..((.(((...((((((.((((......((......))....))))...)))))).))).)).))))))....))))))..... ( -28.60) >DroSec_CAF1 23030 120 + 1 GAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUGGCUGCUUUCUUUUUGCUUUGGCUCAAAGCAAAUUAAUGAUUUGCCAUCAAGCAAAUAAAUCAAAAGCUUCAAAGCAGAUCUCUG ........((.(((((((....(((.......)))))))))).))..(((((((((...))))))))).....(((((((....((((.............))))....))))))).... ( -31.72) >DroSim_CAF1 13250 120 + 1 GAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUGGCUGCUUUCUUUUUGCUUUGGCUCAAAGCAAAUUAAUGAUUUGCCAUCAAGCAAAUAAAUCAAAAGCUUCAAAGCAGAUCUCUG ........((.(((((((....(((.......)))))))))).))..(((((((((...))))))))).....(((((((....((((.............))))....))))))).... ( -31.72) >DroEre_CAF1 22096 120 + 1 GAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUGGCUGCUUUCUUUUUGCUUUGGCUCAAAGCAAAUUAAUGAUUUGCCAUCAAGCAAAUAAAUCAAAAGCUUCAAAGCAGAUCUCUG ........((.(((((((....(((.......)))))))))).))..(((((((((...))))))))).....(((((((....((((.............))))....))))))).... ( -31.72) >consensus GAAAAUCUGACAGCAGCUGAAAAGCAUGCUAAGCUGGCUGCUUUCUUUUUGCUUUGGCUCAAAGCAAAUUAAUGAUUUGCCAUCAAGCAAAUAAAUCAAAAGCUUCAAAGCAGAUCUCUG ....((((...(((((((....))).))))..(((....(((((...(((((((((...)))))))))......((((((......))))))......))))).....)))))))..... (-28.83 = -30.57 + 1.75)



| Location | 22,264,696 – 22,264,816 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -34.21 |
| Energy contribution | -34.02 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22264696 120 - 23771897 CAGAGAUCUGCUUUGAAGCUUUUGAUUUAUUUGCUUGAUGGCAAAUCAUUAAUUUGCUUUGAGCCAGAGCAAAAAGAAAGCACGAAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUC ..(((((((((((((..((((((.....(((((((....)))))))......(((((((((...)))))))))..)))))).)))))).(((((.(((....))))))))..))))))). ( -38.10) >DroSec_CAF1 23030 120 - 1 CAGAGAUCUGCUUUGAAGCUUUUGAUUUAUUUGCUUGAUGGCAAAUCAUUAAUUUGCUUUGAGCCAAAGCAAAAAGAAAGCAGCCAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUC ..(((((((((((((((((..(((((..(((((((....))))))).)))))...)))).....)))))).....((.((((((.(((.......))).....)))))).))))))))). ( -34.90) >DroSim_CAF1 13250 120 - 1 CAGAGAUCUGCUUUGAAGCUUUUGAUUUAUUUGCUUGAUGGCAAAUCAUUAAUUUGCUUUGAGCCAAAGCAAAAAGAAAGCAGCCAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUC ..(((((((((((((((((..(((((..(((((((....))))))).)))))...)))).....)))))).....((.((((((.(((.......))).....)))))).))))))))). ( -34.90) >DroEre_CAF1 22096 120 - 1 CAGAGAUCUGCUUUGAAGCUUUUGAUUUAUUUGCUUGAUGGCAAAUCAUUAAUUUGCUUUGAGCCAAAGCAAAAAGAAAGCAGCCAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUC ..(((((((((((((((((..(((((..(((((((....))))))).)))))...)))).....)))))).....((.((((((.(((.......))).....)))))).))))))))). ( -34.90) >consensus CAGAGAUCUGCUUUGAAGCUUUUGAUUUAUUUGCUUGAUGGCAAAUCAUUAAUUUGCUUUGAGCCAAAGCAAAAAGAAAGCAGCCAGCUUAGCAUGCUUUUCAGCUGCUGUCAGAUUUUC ..((((((((.......((((((.....(((((((....)))))))......(((((((((...)))))))))..))))))........(((((.(((....)))))))).)))))))). (-34.21 = -34.02 + -0.19)


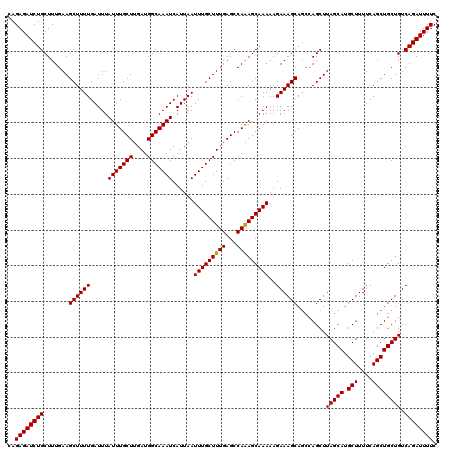
| Location | 22,264,736 – 22,264,850 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -30.74 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22264736 114 + 23771897 CUUUCUUUUUGCUCUGGCUCAAAGCAAAUUAAUGAUUUGCCAUCAAGCAAAUAAAUCAAAAGCUUCAAAGCAGAUCUCUGGCUUAGCGCGCUGCGCUGUGGUGCGGUGCGAUGG- .......((((((.((((......((......))....))))...))))))...(((....(((...((((((....)).)))))))(((((((((....))))))))))))..- ( -33.90) >DroSec_CAF1 23070 115 + 1 CUUUCUUUUUGCUUUGGCUCAAAGCAAAUUAAUGAUUUGCCAUCAAGCAAAUAAAUCAAAAGCUUCAAAGCAGAUCUCUGGCUUAGCGCGCUGCGCUGCGGUGCGGUGCGGUGGG .......(((((((((...))))))))).....(((((((....((((.............))))....))))))).....((((.((((((((((....)))))))))).)))) ( -39.62) >DroEre_CAF1 22136 99 + 1 CUUUCUUUUUGCUUUGGCUCAAAGCAAAUUAAUGAUUUGCCAUCAAGCAAAUAAAUCAAAAGCUUCAAAGCAGAUCUCUGGCUUAGCGCG---------------GUGCGGUGG- .......(((((((((...))))))))).....(((((((....((((.............))))....))))))).......((.(((.---------------..))).)).- ( -20.72) >consensus CUUUCUUUUUGCUUUGGCUCAAAGCAAAUUAAUGAUUUGCCAUCAAGCAAAUAAAUCAAAAGCUUCAAAGCAGAUCUCUGGCUUAGCGCGCUGCGCUG_GGUGCGGUGCGGUGG_ .......(((((((((...)))))))))...........(((((.................(((...((((((....)).)))))))(((((((((....)))))))))))))). (-30.74 = -31.60 + 0.86)

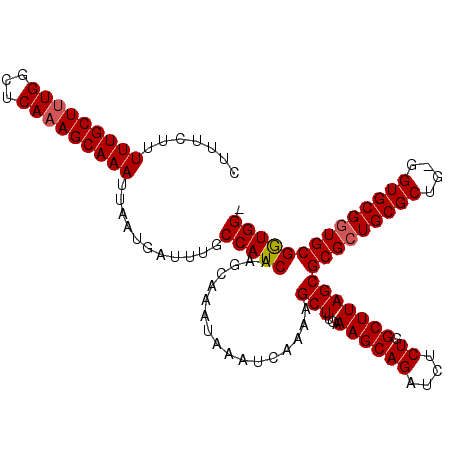
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:45 2006