| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,229,086 – 22,229,208 |
| Length | 122 |
| Max. P | 0.958681 |

| Location | 22,229,086 – 22,229,176 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.91 |
| Mean single sequence MFE | -21.64 |
| Consensus MFE | -17.26 |
| Energy contribution | -18.38 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22229086 90 + 23771897 GUUUCUUCGGUUAUUCGAGGGUUGCGUUGUGGCUAAUACUAAACUUUAAGCAUGAUUAAAGUUUAUAUUAAUUAGUGUGUUCAAUGCUU---G ...(((((((....)))))))..((((((..((((((..((((((((((......)))))))))).....)))))).....))))))..---. ( -22.90) >DroSim_CAF1 6511 93 + 1 GUUUCUUCGGUUAUUCGAGGGUUGCGUUGUGGCUAAUACUAAACUUUAAGCAUGAUUAAAGUUUAUAUUAAUUAGUGUGUUCAAUGCUUGAUG ...(((((((....)))))))..((((((..((((((..((((((((((......)))))))))).....)))))).....))))))...... ( -22.90) >DroEre_CAF1 6494 90 + 1 GUUUCUUCGGUUAUUCGAGGGCUGCGUUGUGGCUAAUACUAAACUUUAAGCAUGAUUAAAGUUUGUAUUAAUUAGUGUAUUCAAUGCUG---G ...(((((((....)))))))..((((((..((((((((.(((((((((......)))))))))))))))......))...))))))..---. ( -23.90) >DroYak_CAF1 8162 90 + 1 GUUUCUUCGGUUAUUCGAGGGUUGCGUUGUAGCUAAUACUAAACUUUAAGCAUGAUUAAAGUUUAUAUUAAUUAGUGUAUUCAAUGCUU---G ...(((((((....)))))))..((((((..((((((..((((((((((......)))))))))).....)))))).....))))))..---. ( -24.00) >DroAna_CAF1 6923 85 + 1 GUUUCUUCGGUUAUUCAAGGGCUGAACUGUGACUAAUACUAAACUU----CACGAUUAAAGUUUAUAUUAAUU-GUGUGUUGAAUGCCU---U ........(((.((((((.(((......))................----(((((((((........))))))-)))).))))))))).---. ( -14.50) >consensus GUUUCUUCGGUUAUUCGAGGGUUGCGUUGUGGCUAAUACUAAACUUUAAGCAUGAUUAAAGUUUAUAUUAAUUAGUGUGUUCAAUGCUU___G ...(((((((....)))))))..((((((..((((((..((((((((((......)))))))))).....)))))).....))))))...... (-17.26 = -18.38 + 1.12)

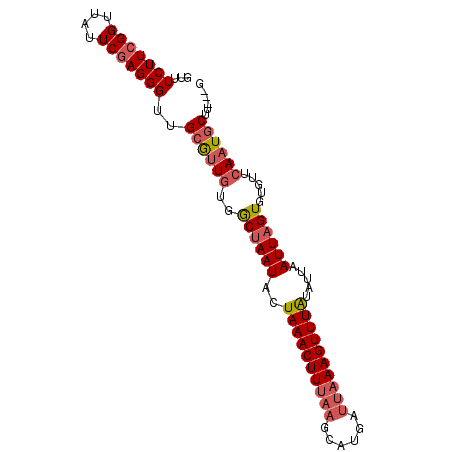

| Location | 22,229,086 – 22,229,176 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 89.91 |
| Mean single sequence MFE | -13.28 |
| Consensus MFE | -9.42 |
| Energy contribution | -9.70 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22229086 90 - 23771897 C---AAGCAUUGAACACACUAAUUAAUAUAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCCACAACGCAACCCUCGAAUAACCGAAGAAAC .---..((.(((......(((((.....((((((((((......))))))))))..)))))...))).)).....(((......)))...... ( -13.00) >DroSim_CAF1 6511 93 - 1 CAUCAAGCAUUGAACACACUAAUUAAUAUAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCCACAACGCAACCCUCGAAUAACCGAAGAAAC ..((..((.(((......(((((.....((((((((((......))))))))))..)))))...))).)).....(((......))).))... ( -13.30) >DroEre_CAF1 6494 90 - 1 C---CAGCAUUGAAUACACUAAUUAAUACAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCCACAACGCAGCCCUCGAAUAACCGAAGAAAC .---..((...............(((((((((((((((......))))))))).))))))((......)).))..(((......)))...... ( -14.40) >DroYak_CAF1 8162 90 - 1 C---AAGCAUUGAAUACACUAAUUAAUAUAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCUACAACGCAACCCUCGAAUAACCGAAGAAAC .---..((.(((......(((((.....((((((((((......))))))))))..)))))...))).)).....(((......)))...... ( -13.00) >DroAna_CAF1 6923 85 - 1 A---AGGCAUUCAACACAC-AAUUAAUAUAAACUUUAAUCGUG----AAGUUUAGUAUUAGUCACAGUUCAGCCCUUGAAUAACCGAAGAAAC .---.(((....(((....-.((((((((((((((((....))----)))))).))))))))....)))..)))................... ( -12.70) >consensus C___AAGCAUUGAACACACUAAUUAAUAUAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCCACAACGCAACCCUCGAAUAACCGAAGAAAC ......................((((((((((((((((......))))))))).)))))))..............(((......)))...... ( -9.42 = -9.70 + 0.28)

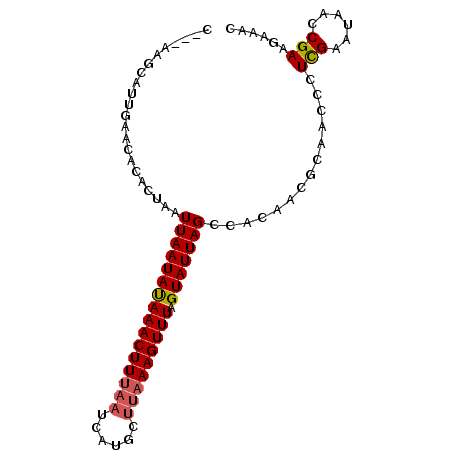

| Location | 22,229,101 – 22,229,208 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Mean single sequence MFE | -12.75 |
| Consensus MFE | -8.26 |
| Energy contribution | -8.65 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22229101 107 - 23771897 AAAA--UAAGACAACAACA------AAAACAAAGGAAAAGC---AAGCAUUGAACACACUAAUUAAUAUAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCCACAACGCAACCCUCG ....--.............------................---..((.(((......(((((.....((((((((((......))))))))))..)))))...))).))........ ( -11.70) >DroGri_CAF1 6906 103 - 1 AAACAUCACGACAC--------AACAACACACAGAAUACACUUCA---AUUUAA----CUCAUUAAUAUAAACUUUAAUCGCGUUUAAAGUUGAAUAUUAGUCACAAAGCAAACCCAC .........(((..--------...........(((.....))).---......----.....((((((.((((((((......))))))))..)))))))))............... ( -12.40) >DroSim_CAF1 6526 116 - 1 AAAA--UAAGACAACAAAAAAAAAAAAAACAAAGGACAAGCAUCAAGCAUUGAACACACUAAUUAAUAUAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCCACAACGCAACCCUCG ....--..........................(((.(((((.....)).)))......(((((.....((((((((((......))))))))))..)))))............))).. ( -13.60) >DroEre_CAF1 6509 102 - 1 AAAA--UAAGACAA-----------GAUACAAAGGAAAAGC---CAGCAUUGAAUACACUAAUUAAUACAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCCACAACGCAGCCCUCG ....--........-----------........((.....)---).((...............(((((((((((((((......))))))))).))))))((......)).))..... ( -14.00) >DroYak_CAF1 8177 102 - 1 AAAA--UAAGACAA-----------AAAACAAAGGAAAAGC---AAGCAUUGAAUACACUAAUUAAUAUAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCUACAACGCAACCCUCG ....--........-----------.......(((......---...................(((((((((((((((......))))))))).))))))((......))...))).. ( -12.10) >DroAna_CAF1 6938 93 - 1 AAAC------ACAC-----------AAAACAUUUAAAAAAA---AGGCAUUCAACACAC-AAUUAAUAUAAACUUUAAUCGUG----AAGUUUAGUAUUAGUCACAGUUCAGCCCUUG ....------....-----------................---.(((....(((....-.((((((((((((((((....))----)))))).))))))))....)))..))).... ( -12.70) >consensus AAAA__UAAGACAA___________AAAACAAAGGAAAAGC___AAGCAUUGAACACACUAAUUAAUAUAAACUUUAAUCAUGCUUAAAGUUUAGUAUUAGCCACAACGCAACCCUCG ..............................................................((((((((((((((((......))))))))).)))))))................. ( -8.26 = -8.65 + 0.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:38 2006