| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,214,859 – 22,214,961 |
| Length | 102 |
| Max. P | 0.967913 |

| Location | 22,214,859 – 22,214,961 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.43 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -12.48 |
| Energy contribution | -14.20 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.32 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
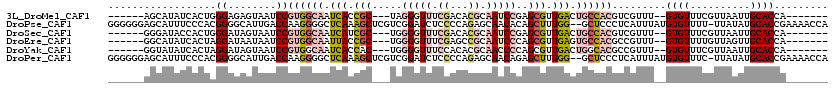
>3L_DroMel_CAF1 22214859 102 + 23771897 ------AGCAUAUCACUGGGAGAGUAAUCCGUGGCAAUCACCGC---UAGGGUUUCGACACGCAAUCCGAGCGUUGACUGCCACGUCGUUU--GUGUUUCGUUAAUUGCACCA------- ------.(((....((.(..(.(..(((.(((((((.(((.(((---(.(((((.((...)).))))).)))).))).)))))))..))).--.).)..)))....)))....------- ( -33.20) >DroPse_CAF1 12751 117 + 1 GGGGGGAGCAUUUCCCACGGGGCAUUGACCAAGGGGCUCAAAGCUCGUCGGAUCUCCCCAGAGCAACACAGCUUUGG--GCUCCCUCAUUUAUGUGUUUU-UUAUAUGCACCGAAAACCA .((((((((...(((.(((((.(.((((((....)).)))).)))))).))).....(((((((......)))))))--))))))))......((((...-......))))......... ( -43.90) >DroSec_CAF1 11842 102 + 1 ------GGGAUACCACUGGGAUAGUAAUCCGUGGCAAUCAUCGC---UGGGGUUUCGACACGCAAUCCGAGCGUUGACUGCCACGUCGUUU--GUGUUUCGUUAAUUGCACCA------- ------(..((((.....((((....))))((((((.(((.(((---(.(((((.((...)).))))).)))).))).)))))).......--))))..).............------- ( -37.90) >DroEre_CAF1 11589 102 + 1 ------GGCAUAUCACUAGGAUAAUAAUCCGUGGCAAUUACCGC---UGGGGUUUCGAGCCGCAAUCCCAGCGUUGAGUGCCACGCCGUUU--GUGUUUUGUUAGUUGCACCA------- ------(((.........((((....))))((((((.(((.(((---(((((((.((...)).)))))))))).))).)))))))))....--((((..(....)..))))..------- ( -42.00) >DroYak_CAF1 10848 102 + 1 ------GGUAUAUCACUAGGAUAGUAAUCCGUGGCAAUCACCAC---UGGGGUUUCCACACGCAACCCCAGCGUUGACUGGCACGCCGUUU--GUGUUUCGUUAAUUGCACCA------- ------(((.........((((....))))(((.((.(((.(.(---(((((((.(.....).)))))))).).))).)).))))))....--((((..........))))..------- ( -32.90) >DroPer_CAF1 13043 117 + 1 GGGGGGAGCAUUUCCCACGGGGCAUUGACCAAGGGGCUCAAAGCUCGUCGGAUCUCCCCAGAGCAACAGAGCUUUGG--GCUCCCUCAUUUAUGUGUUUC-UUAUAUGCACCGAAAACCA .((((((((...(((.(((((.(.((((((....)).)))).)))))).))).....(((((((......)))))))--))))))))......((((...-......))))......... ( -43.90) >consensus ______AGCAUAUCACUAGGAGAAUAAUCCGUGGCAAUCACCGC___UGGGGUUUCCACACGCAAUCCCAGCGUUGACUGCCACGUCGUUU__GUGUUUCGUUAAUUGCACCA_______ ..................((........))((((((.(((.(((.....(((((.((...)).)))))..))).))).)))))).........((((..........))))......... (-12.48 = -14.20 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:35 2006