
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,201,649 – 22,201,808 |
| Length | 159 |
| Max. P | 0.880635 |

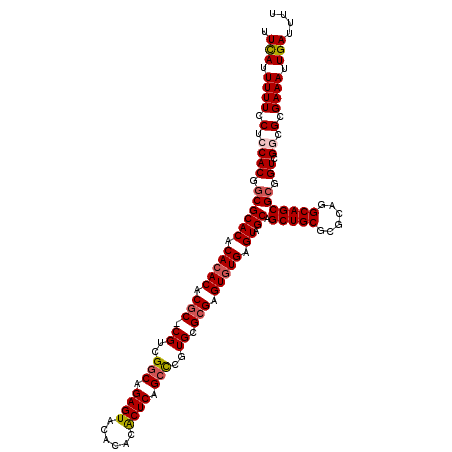
| Location | 22,201,649 – 22,201,768 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -47.58 |
| Consensus MFE | -38.80 |
| Energy contribution | -39.52 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22201649 119 + 23771897 UUCAUUUUUCCUCCACGGCGCACACACACGCGC-CGUCGGCAGAGUACACACACUCAGCCCGUGCGCGAGUGUGAGUAGCAGCUGCGCGCAGGCAGCGCGGUCGGCGCGAAAUUGAUUUU .(((..(((((.((((.((((.(((((.(((((-((..(((.((((......)))).))))).))))).))))).))....(((((......))))))).)).)).).)))).))).... ( -49.60) >DroSec_CAF1 60581 119 + 1 UUCAUUUUUCCUCCACGGCGCACACACACACGC-CGUCGGCAGAGUACACACACUCAGCUCGUGCGCGAGUGUGAGUGGCAGCUGCGCGCAGGCAGCGCGGUCGGCGCGAAAUUGAUUUU .(((..((((((((((((((..........)))-))).)).))......(((((((.((....))..))))))).(((.(.(((((((.......))))))).).))))))).))).... ( -46.60) >DroSim_CAF1 56238 119 + 1 UUCAUUUUUCCUCCACGGCGCACACACACACGC-CGUCGGCAGAGUACACACACUCAGCCCGUGCGCGAGUGUGAGUGGCAGCUGCGCGCAGGCAGCGCGGUCGGCGCGAAAUUGAUUUU .(((..(((((.((((.((((((.(((((.(((-((..(((.((((......)))).)))..)).))).))))).)).)).(((((......))))))).)).)).).)))).))).... ( -47.70) >DroEre_CAF1 60380 120 + 1 UUUAGUUUUCCCACACGCCGCACACACACACACCCGUCGACAGAGUACACACACUCAGCCCGUGCGCGGGUGUGAGUAGCAGCUGCGCGCAGGCAGCGCCGUCGGCGCGAAAUUGAUUUU .((((((((......(((((.((..((.(((((((((.....((((......)))).((....))))))))))).)).((.(((((......))))))).))))))).)))))))).... ( -43.90) >DroYak_CAF1 72109 119 + 1 UUCAUUUUUCCUACACGGCGCACACACACUCGC-CGUGGGCAGAGUACACACGCUCAGCCCGUGCGCGAGUAUGAGUAGCAGCUGCGCGCAAGCAGCGCUGUCGGCGCGAAAUUGAUUUU .(((..(((((..((((((((((.((.((((((-(((((((.((((......)))).))))))).)))))).)).)).)).(((((......)))))))))).)..).)))).))).... ( -50.10) >consensus UUCAUUUUUCCUCCACGGCGCACACACACACGC_CGUCGGCAGAGUACACACACUCAGCCCGUGCGCGAGUGUGAGUAGCAGCUGCGCGCAGGCAGCGCGGUCGGCGCGAAAUUGAUUUU .(((.((((.(.((((.((((((.(((((.(((.((..(((.((((......)))).)))..)).))).))))).)).)).(((((......))))))).)).)).).)))).))).... (-38.80 = -39.52 + 0.72)

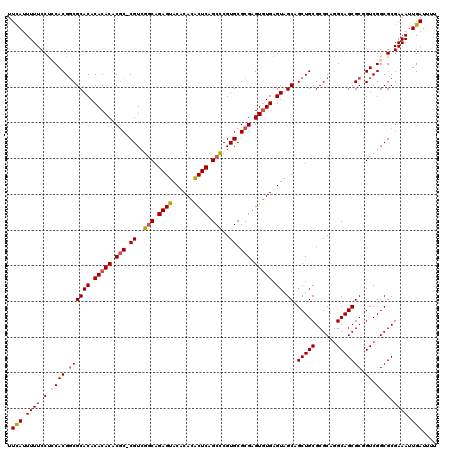
| Location | 22,201,649 – 22,201,768 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -48.96 |
| Consensus MFE | -41.58 |
| Energy contribution | -41.46 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22201649 119 - 23771897 AAAAUCAAUUUCGCGCCGACCGCGCUGCCUGCGCGCAGCUGCUACUCACACUCGCGCACGGGCUGAGUGUGUGUACUCUGCCGACG-GCGCGUGUGUGUGCGCCGUGGAGGAAAAAUGAA ....(((.....((((.....))))..(((.((((..((.((....(((((.(((((.(((((.(((((....))))).)))..))-))))).))))).)))))))).))).....))). ( -50.20) >DroSec_CAF1 60581 119 - 1 AAAAUCAAUUUCGCGCCGACCGCGCUGCCUGCGCGCAGCUGCCACUCACACUCGCGCACGAGCUGAGUGUGUGUACUCUGCCGACG-GCGUGUGUGUGUGCGCCGUGGAGGAAAAAUGAA ........(((((((.(((((((((((((..((.((((..(..((.((((((((.((....)))))))))).)).).))))))..)-))).))))).)).)).))))))).......... ( -46.40) >DroSim_CAF1 56238 119 - 1 AAAAUCAAUUUCGCGCCGACCGCGCUGCCUGCGCGCAGCUGCCACUCACACUCGCGCACGGGCUGAGUGUGUGUACUCUGCCGACG-GCGUGUGUGUGUGCGCCGUGGAGGAAAAAUGAA ....(((.....((((.....))))..(((.((((..((.((....(((((.(((((.(((((.(((((....))))).)))..))-))))).))))).)))))))).))).....))). ( -47.60) >DroEre_CAF1 60380 120 - 1 AAAAUCAAUUUCGCGCCGACGGCGCUGCCUGCGCGCAGCUGCUACUCACACCCGCGCACGGGCUGAGUGUGUGUACUCUGUCGACGGGUGUGUGUGUGUGCGGCGUGUGGGAAAACUAAA ............(((((...)))))..((..(((((.((.(((((.((((((((((.(((....(((((....)))))))))).)))))))).))).)))).)))))..))......... ( -52.10) >DroYak_CAF1 72109 119 - 1 AAAAUCAAUUUCGCGCCGACAGCGCUGCUUGCGCGCAGCUGCUACUCAUACUCGCGCACGGGCUGAGCGUGUGUACUCUGCCCACG-GCGAGUGUGUGUGCGCCGUGUAGGAAAAAUGAA ....(((.....((((.....))))..((((((((..((.((.((.(((((((((.(..((((.(((........))).))))..)-))))))))).)))))))))))))).....))). ( -48.50) >consensus AAAAUCAAUUUCGCGCCGACCGCGCUGCCUGCGCGCAGCUGCUACUCACACUCGCGCACGGGCUGAGUGUGUGUACUCUGCCGACG_GCGUGUGUGUGUGCGCCGUGGAGGAAAAAUGAA ............((((.....))))..(((.((((..((.((....(((((.(((((.(((((.(((((....))))).)))..)).))))).))))).)))))))).)))......... (-41.58 = -41.46 + -0.12)


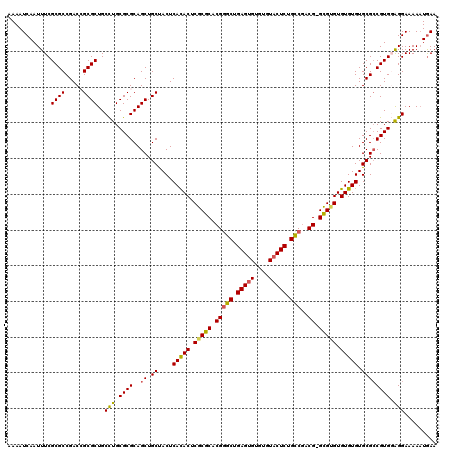
| Location | 22,201,688 – 22,201,808 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -46.06 |
| Consensus MFE | -42.46 |
| Energy contribution | -41.62 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22201688 120 + 23771897 CAGAGUACACACACUCAGCCCGUGCGCGAGUGUGAGUAGCAGCUGCGCGCAGGCAGCGCGGUCGGCGCGAAAUUGAUUUUUCGCGGCAGAAGUUUUCUCGAGCCAGCAGGCACUUUCCUU ..((((((.(((((((.((....))..))))))).)))((.(((((((((.....)))))..))))((((((......)))))).)).........)))..(((....)))......... ( -45.30) >DroSec_CAF1 60620 120 + 1 CAGAGUACACACACUCAGCUCGUGCGCGAGUGUGAGUGGCAGCUGCGCGCAGGCAGCGCGGUCGGCGCGAAAUUGAUUUUUCGCGGCAGAAGUUUUCUUGAGCCAGCAGGCAUUUUCCUU .....(((.(((((((.((....))..))))))).)))((.(((((......))))))).(((.((((((((......))))))(((..(((....)))..))).)).)))......... ( -46.40) >DroSim_CAF1 56277 120 + 1 CAGAGUACACACACUCAGCCCGUGCGCGAGUGUGAGUGGCAGCUGCGCGCAGGCAGCGCGGUCGGCGCGAAAUUGAUUUUUCGCGGCAGAAGUUUUCUUGAGCCAGCAGGCACUUUCCUU .........(((((((.((....))..)))))))((((.(.(((((((.......)))))))..((((((((......))))))(((..(((....)))..))).)).).))))...... ( -46.20) >DroEre_CAF1 60420 120 + 1 CAGAGUACACACACUCAGCCCGUGCGCGGGUGUGAGUAGCAGCUGCGCGCAGGCAGCGCCGUCGGCGCGAAAUUGAUUUUUCGCGGCAGAAGUUUUCUCGAGCUAGCAGGCAUUUUCCCU ..((((......)))).(((.((((((((.(((.....))).)))))))).)))...(((...(.(((((((......))))))).)...(((((....)))))....)))......... ( -48.80) >DroYak_CAF1 72148 120 + 1 CAGAGUACACACGCUCAGCCCGUGCGCGAGUAUGAGUAGCAGCUGCGCGCAAGCAGCGCUGUCGGCGCGAAAUUGAUUUUUCGCGGCAGAAGUUUUCUUGAGCUAGUAGGCAUUUUCCUU ..((((......)))).((((((((....)))))(.((((.(((((((....)).((((.....))))((((......)))))))))..(((....)))..)))).).)))......... ( -43.60) >consensus CAGAGUACACACACUCAGCCCGUGCGCGAGUGUGAGUAGCAGCUGCGCGCAGGCAGCGCGGUCGGCGCGAAAUUGAUUUUUCGCGGCAGAAGUUUUCUUGAGCCAGCAGGCAUUUUCCUU ..((((((.(((((((.((....))..))))))).)))((..((((((((.....))))......(((((((......)))))))))))..))...)))..(((....)))......... (-42.46 = -41.62 + -0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:30 2006