| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,165,669 – 22,165,775 |
| Length | 106 |
| Max. P | 0.678629 |

| Location | 22,165,669 – 22,165,775 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.59 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -18.05 |
| Energy contribution | -18.38 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
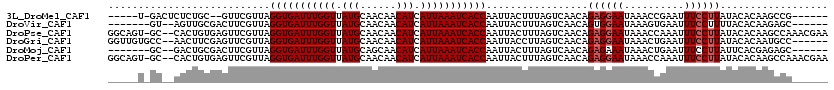
>3L_DroMel_CAF1 22165669 106 - 23771897 -----U-GACUCUCUGC--GUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUACACAAGCCG------ -----(-((((....((--....))..(((((((((((.(((......))).))))))))))).........)))))...((((((..........))))))............------ ( -23.00) >DroVir_CAF1 49694 105 - 1 -------GU--AGUUGCGACUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGUGGAAUAAAGUGAAUUUCCUUUUACACAAGAGC------ -------((--((..(.((........(((((((((((.(((......))).)))))))))))..((((((((.((.......)).))))))))...)))..))))........------ ( -24.60) >DroPse_CAF1 48375 117 - 1 GGCAGU-GC--CACUGUGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCAAAUUUCCUUAUACACAAGCCAAACGAA (((.((-..--.((((.((((..(((.(((((((((((.(((......))).)))))))))))))).))))))))..)).((((((..........))))))........)))....... ( -30.30) >DroGri_CAF1 27082 112 - 1 GGUUGUGCC--AACUUCGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACCUUAGUCAACAGAGGAAUAAACUGAAUUUCCUUAUACACAAUGCC------ .((((((..--......(((((((((.(((((((((((.(((......))).))))))))))).....((((.(....).))))....))).)))))).......))))))...------ ( -27.76) >DroMoj_CAF1 77546 105 - 1 -------GC--GACUGCGACUUCGUUAGGUGAUUUGGUUAUGCAGCAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGAAAUAAACUGAAUUUCCUUAUUCACGAGAGC------ -------((--(((((((....)))..(((((((((((.(((......))).))))))))))).........))))...............(((((......))))).....))------ ( -23.30) >DroPer_CAF1 48379 117 - 1 GGCAGU-GC--CACUGUGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCAAAUUUCCUUAUACACAAGCCAAACGAA (((.((-..--.((((.((((..(((.(((((((((((.(((......))).)))))))))))))).))))))))..)).((((((..........))))))........)))....... ( -30.30) >consensus _____U_GC__CACUGCGAGUUCGUUAGGUGAUUUGGUUAUGCAACAACAUCAUUAAAUCACCAAUUACUUUAGUCAACAGAGGAAUAAACCGAAUUUCCUUAUACACAAGCCC______ ...........................(((((((((((.(((......))).))))))))))).................((((((..........)))))).................. (-18.05 = -18.38 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:22 2006