
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,153,714 – 22,153,822 |
| Length | 108 |
| Max. P | 0.505552 |

| Location | 22,153,714 – 22,153,822 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.36 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -15.85 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
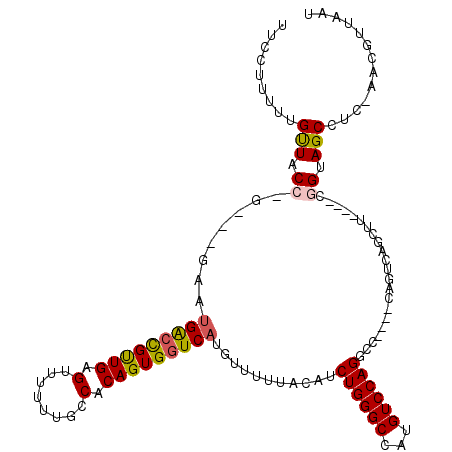
>3L_DroMel_CAF1 22153714 108 + 23771897 UUC-UUUGUGUUUCCGGG--AACUGACCGUUGAGUUUUUUGCCACAGUGGUCAUGUUUUUACAUCUGGGCCAUGUCCAGGCC----CAGUCAGCUU----CGGUAGCCUC-GGCGUUAAU ...-.........(((((--((((((((((((.((.....))..))))))))).))).......(((((((.......))))----)))...((..----..))...)))-))....... ( -32.00) >DroVir_CAF1 36436 94 + 1 -UCAUUUUUGCUUCC-----GAAUGACUGUUGAGUUUUUUUUCACAGUGGUCAUGUUUUUACAACUGGGCCAUGUCCAG-----------CAGCGC----C-GUAGCC----ACGUUAAU -........(((..(-----(.(((((..(((.(........).)))..)))))..........((((((...))))))-----------......----)-).))).----........ ( -19.30) >DroPse_CAF1 35473 118 + 1 UUUCUU--UGUUCCCUGACUGACUGACCGUUGGGUUUUUUGCCACAGUGGUCAUGUUUUUACAUCUGGGCCAUGUCCAGGCCCCCCCAGUCAGCUUCGUUCGGUAGCCUCCAACGUUAAU ......--......(((((((((((((((((((((.....))).))))))))).))..........(((((.......)))))...)))))))...((((.((......))))))..... ( -37.90) >DroGri_CAF1 13704 93 + 1 -GCUUUUUAGUUACU-----GAAUGACUGCUGAGUUUU-UUUCACAGUGGUCAUGUUUUUACAACUGGGCCAUGUCCAG-----------CAACGA----C-GUAGCC----ACGUUAAU -((((..((((((..-----...))))))..))))...-.......((((..(((((.((....((((((...))))))-----------.)).))----)-))..))----))...... ( -23.80) >DroAna_CAF1 11783 109 + 1 CUCCCUUUCGCUACCGGG--CAAGGGGCGUUGAGUUUUUUGCCACAGUGGUCGUGUUUUUACAUCUGGGCCAUGUCCAGGCC----CAGUCAGCUU----CGGUAGCCUC-AGCGUUAAU .........(((((((((--((((((((.....))))))))))..(((....((((....))))(((((((.......))))----)))...))).----)))))))...-......... ( -40.20) >DroPer_CAF1 35513 118 + 1 UUUCUU--UGUUCCCUGACUGACUGACCGUUGGGUUUUUUGCCACAGUGGUCAUGUUUUUACAUCUGGGCCAUGUCCAGGCCCCCCCAGUCAGCUUCGUUCGGUAGCCUCCAACGUUAAU ......--......(((((((((((((((((((((.....))).))))))))).))..........(((((.......)))))...)))))))...((((.((......))))))..... ( -37.90) >consensus UUCCUUUUUGUUACC_G___GAAUGACCGUUGAGUUUUUUGCCACAGUGGUCAUGUUUUUACAUCUGGGCCAUGUCCAGGCC____CAGUCAGCUU____CGGUAGCCUC_AACGUUAAU .........(((.((........(((((((((.(........).)))))))))...........((((((...))))))......................)).)))............. (-15.85 = -15.80 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:17 2006