| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,150,340 – 22,150,469 |
| Length | 129 |
| Max. P | 0.999104 |

| Location | 22,150,340 – 22,150,443 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -23.59 |
| Energy contribution | -23.48 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22150340 103 + 23771897 UCUGGGGAGAAACCAUAUUAGAUUUGUUAAUUUAAAUGCAAUUGCCAAAAGCAGUUGCACCUUGAGCCAAAUGGCCUUUGCCUUCCUACCAUAAAUAAAUCAA ..(((((((........(((((((....))))))).(((((((((.....)))))))))......(((....)))......)))))))............... ( -25.30) >DroSec_CAF1 9779 103 + 1 UCUGGGGAGAAACCAUAUUAGAUUUGUUAAUUUAAAUGCAAUUGCCAAAAGCAGUUGCACCUUGAGCCCAAUGGCCUUUACCUUUCUACCAUAAAUAAAUCAA ..(((.......))).....((((((((...((((.(((((((((.....)))))))))..))))(((....)))..................)))))))).. ( -23.30) >DroSim_CAF1 9621 103 + 1 UCUGGGGAGAAACCAUAUUAGAUUUGUUAAUUUAAAUGCAAUUGCCAAAAGCAGUUGCACCUUGAGCCAAAUGGCCUUUGCCUUCCUACCAUAAAUAAAUCAA ..(((((((........(((((((....))))))).(((((((((.....)))))))))......(((....)))......)))))))............... ( -25.30) >DroEre_CAF1 9666 103 + 1 UCUGGGGAGAAACCAUAUUAGAUUUGUUAAUUUAAAUGCAAUUGCCGAAAGCAGUUGCACCUUGAGCCAAAUGGCCUUUGCCUUCGUACCAUAAAUAAAUCAA ..(((.......))).....((((((((...((((.((((((((.(....)))))))))..))))(((....)))..................)))))))).. ( -24.70) >DroYak_CAF1 9375 103 + 1 UCUGGGGAGAAACCAUAUUAGAUUUGUUAAUUUAAAUGCAAUUGCCAAAAGCAGUUGCACCUUGAGCCAAAUGGCCUUUGCCUUCCUACCAUAAAUAAAUCAA ..(((((((........(((((((....))))))).(((((((((.....)))))))))......(((....)))......)))))))............... ( -25.30) >DroAna_CAF1 8718 99 + 1 UCGGAGGAGAAACCUCAUUAGAUUUGUUAAUUUAAAUGCAAUUGCCGGAGGCAGUUGCACCUUGAGCCGGCCGGCCUUUGCCUUC----CAUAAAUAAAUCAA ...((((.....))))....((((((((..(((((.((((((((((...))))))))))..)))))..((..(((....)))..)----)...)))))))).. ( -31.90) >consensus UCUGGGGAGAAACCAUAUUAGAUUUGUUAAUUUAAAUGCAAUUGCCAAAAGCAGUUGCACCUUGAGCCAAAUGGCCUUUGCCUUCCUACCAUAAAUAAAUCAA ..(((.......))).....((((((((...((((.(((((((((.....)))))))))..))))(((....)))..................)))))))).. (-23.59 = -23.48 + -0.11)

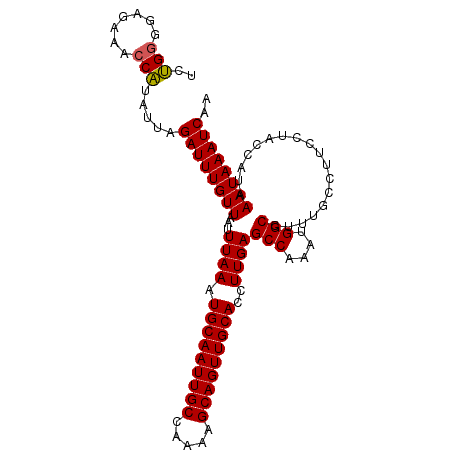

| Location | 22,150,340 – 22,150,443 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22150340 103 - 23771897 UUGAUUUAUUUAUGGUAGGAAGGCAAAGGCCAUUUGGCUCAAGGUGCAACUGCUUUUGGCAAUUGCAUUUAAAUUAACAAAUCUAAUAUGGUUUCUCCCCAGA ............(((..(((((.((..((((....))))..((((((((.(((.....))).))))))))..................)).)))))..))).. ( -27.10) >DroSec_CAF1 9779 103 - 1 UUGAUUUAUUUAUGGUAGAAAGGUAAAGGCCAUUGGGCUCAAGGUGCAACUGCUUUUGGCAAUUGCAUUUAAAUUAACAAAUCUAAUAUGGUUUCUCCCCAGA ....(((((.....)))))..(((....))).(((((....((((((((.(((.....))).))))))))........(((((......)))))...))))). ( -21.60) >DroSim_CAF1 9621 103 - 1 UUGAUUUAUUUAUGGUAGGAAGGCAAAGGCCAUUUGGCUCAAGGUGCAACUGCUUUUGGCAAUUGCAUUUAAAUUAACAAAUCUAAUAUGGUUUCUCCCCAGA ............(((..(((((.((..((((....))))..((((((((.(((.....))).))))))))..................)).)))))..))).. ( -27.10) >DroEre_CAF1 9666 103 - 1 UUGAUUUAUUUAUGGUACGAAGGCAAAGGCCAUUUGGCUCAAGGUGCAACUGCUUUCGGCAAUUGCAUUUAAAUUAACAAAUCUAAUAUGGUUUCUCCCCAGA ............(((...((((.((..((((....))))..((((((((.(((.....))).))))))))..................)).))))...))).. ( -23.40) >DroYak_CAF1 9375 103 - 1 UUGAUUUAUUUAUGGUAGGAAGGCAAAGGCCAUUUGGCUCAAGGUGCAACUGCUUUUGGCAAUUGCAUUUAAAUUAACAAAUCUAAUAUGGUUUCUCCCCAGA ............(((..(((((.((..((((....))))..((((((((.(((.....))).))))))))..................)).)))))..))).. ( -27.10) >DroAna_CAF1 8718 99 - 1 UUGAUUUAUUUAUG----GAAGGCAAAGGCCGGCCGGCUCAAGGUGCAACUGCCUCCGGCAAUUGCAUUUAAAUUAACAAAUCUAAUGAGGUUUCUCCUCCGA ..(((((.....((----(..(((....)))..))).....((((((((.((((...)))).))))))))........)))))....((((.....))))... ( -28.10) >consensus UUGAUUUAUUUAUGGUAGGAAGGCAAAGGCCAUUUGGCUCAAGGUGCAACUGCUUUUGGCAAUUGCAUUUAAAUUAACAAAUCUAAUAUGGUUUCUCCCCAGA ............(((..(((((.((..((((....))))..((((((((.(((.....))).))))))))..................)).)))))..))).. (-22.04 = -22.90 + 0.86)


| Location | 22,150,374 – 22,150,469 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -13.15 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22150374 95 + 23771897 AAUGCAAUUGCCAAAAGCAGUUGCACCUUGAG-----------CCAAAUGGCCUUUGCCUUCCUACCAUAAAUAAAUCAAAUUAAUGUCAAUUGCACUGAAAACAA-------- ..(((((((((.....)))))))))..(((((-----------((....)))..............(((.(((.......))).)))))))...............-------- ( -16.10) >DroSim_CAF1 9655 97 + 1 AAUGCAAUUGCCAAAAGCAGUUGCACCUUGAG-----------CCAAAUGGCCUUUGCCUUCCUACCAUAAAUAAAUCAAAUUAAUGUCAAUUGCACCGACAACAAC------A ..(((((((((.....)))))))))..(((.(-----------((....))).................................((((.........)))).))).------. ( -18.10) >DroEre_CAF1 9700 100 + 1 AAUGCAAUUGCCGAAAGCAGUUGCACCUUGAG-----------CCAAAUGGCCUUUGCCUUCGUACCAUAAAUAAAUCAAAUUAAUGUCAAUUGCACCGAACACAACA---ACA ..((((((((.((((.((((..((...(((..-----------.)))...))..)))).))))...(((.(((.......))).))).))))))))............---... ( -17.50) >DroYak_CAF1 9409 99 + 1 AAUGCAAUUGCCAAAAGCAGUUGCACCUUGAG-----------CCAAAUGGCCUUUGCCUUCCUACCAUAAAUAAAUCAAAUUAAUGUCAAUUGCACCGAAAACAACA---AC- ..(((((((((.....)))))))))..(((((-----------((....)))..............(((.(((.......))).))))))).................---..- ( -16.10) >DroAna_CAF1 8752 92 + 1 AAUGCAAUUGCCGGAGGCAGUUGCACCUUGAG-----------CCGGCCGGCCUUUGCCUUC----CAUAAAUAAAUCAAAUUAGUGUCAAUUGCACGAACAACAAG------- ..((((((((((...)))))))))).((((..-----------..((..(((....)))..)----).................((((.....))))......))))------- ( -25.40) >DroPer_CAF1 32338 107 + 1 CCAGCAAAUCCUGGAAGCACUUGCACCUUGAGGAGCUAAAAUGGCCAAUGGCCUUUGCCAUC-UGCAAUAAAUAAAUCAAAUUAAUGUCAAUUGCC---ACAACAAA-UGA--A ((((......))))..(((.(((.((.((((.((.........((..(((((....))))).-.))..........))...)))).))))).))).---........-...--. ( -20.51) >consensus AAUGCAAUUGCCAAAAGCAGUUGCACCUUGAG___________CCAAAUGGCCUUUGCCUUCCUACCAUAAAUAAAUCAAAUUAAUGUCAAUUGCACCGAAAACAAC_______ ..((((((((.((...((....)).........................(((....)))..........................)).)))))))).................. (-13.15 = -13.27 + 0.11)
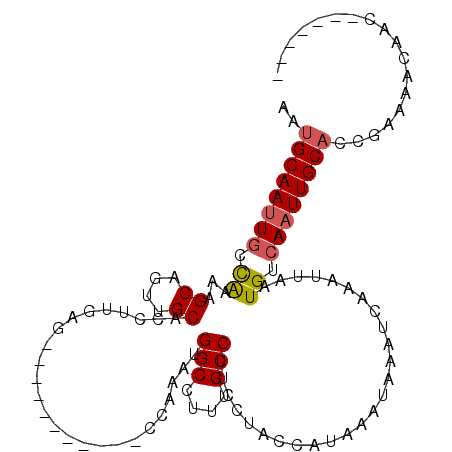
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:16 2006