| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,139,198 – 22,139,304 |
| Length | 106 |
| Max. P | 0.929421 |

| Location | 22,139,198 – 22,139,304 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.68 |
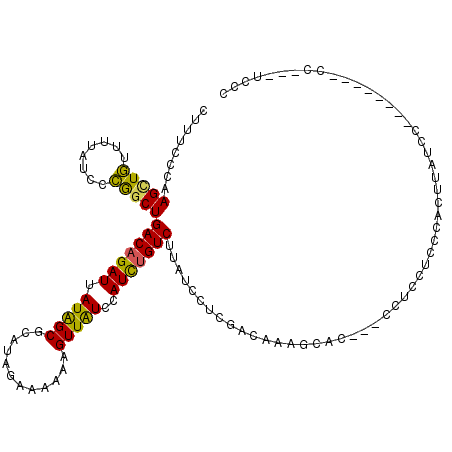
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -14.14 |
| Energy contribution | -13.45 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
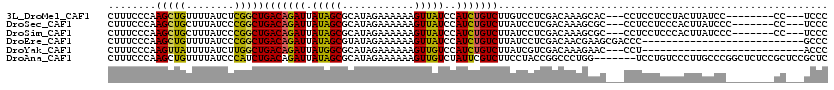
>3L_DroMel_CAF1 22139198 106 - 23771897 CUUUCCCAAGCUGUUUUAUCUCGGCUGACAGAUUAUAGCGCAUAGAAAAAAGUUAUCCAUCUGUCUUGUCCUCGACAAAGCAC---CCUCCUCCUACUUAUCC--------CC---UCCC ........(((((........)))))(((((((.(((((............)))))..)))))))(((((...))))).....---.................--------..---.... ( -18.50) >DroSec_CAF1 125035 107 - 1 CUUUCCCAAGCUGCUUUAUCCCGGCUGACAGAUUAUAGCGCAUAGAAAAAAGUUAUCCAUCUGUCUUAUCCUCGACAAAGCGC---CCUCCUCCCACUUAUCCC-------CC---UCCC .........((.(((((.((..((..(((((((.(((((............)))))..)))))))....))..)).)))))))---..................-------..---.... ( -20.10) >DroSim_CAF1 133931 107 - 1 CUUUCCCAAGCUGCUUUAUCCCGGCUGACAGAUUAUAGCGCAUAGAAAAAAGUUAUCCAUCUGUCUUAUCCUCGACAAAGCGC---CCUCCUCCCACUUAUCCC-------CC---UCCC .........((.(((((.((..((..(((((((.(((((............)))))..)))))))....))..)).)))))))---..................-------..---.... ( -20.10) >DroEre_CAF1 124688 93 - 1 CUUUCCCAAGCUGUUUUAUCCCGGCUGACAGAUUAUAGCGUAUAGAAAAAAGUUAUCCAUCUGUCUUAUCCUCGACAACGAAGCGACCC---------------------------GCCC ........(((((........)))))(((((((.(((((............)))))..)))))))......(((....))).(((...)---------------------------)).. ( -17.50) >DroYak_CAF1 173295 90 - 1 CUUUCCCAAGUUAUUUUAUCUUGGCUGACAGAUUAUGGCGCAUAGAAAAAAGUUGUCCAUCUGUCUUAUCGUCGACAAAGAAC---CCU---------------------------ACCC ((((.(((((.........)))))..((((((...(((.(((...........))))))))))))...........))))...---...---------------------------.... ( -14.90) >DroAna_CAF1 141323 113 - 1 CUUUCCCAAGCUGUUUUAUCCCAUCUGACAGAUUAUAGCGCAUAGAAAAAAGUUGUCUAUUCGUCUUCCUACCGGCCCUGG-------UCCUGUCCCUUGCCCGGCUCUCCGCUCCGCUC ..........................(((((......(((.(((((.........))))).))).........(((....)-------)))))))....((..(((.....)))..)).. ( -17.00) >consensus CUUUCCCAAGCUGUUUUAUCCCGGCUGACAGAUUAUAGCGCAUAGAAAAAAGUUAUCCAUCUGUCUUAUCCUCGACAAAGCAC___CCUCCUCCCACUUAUCC________CC___UCCC ........(((((........)))))(((((((.(((((............)))))..)))))))....................................................... (-14.14 = -13.45 + -0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:10 2006