
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
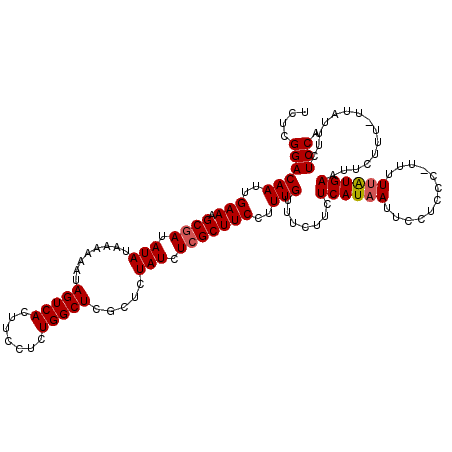
| Location | 22,133,871 – 22,133,990 |
| Length | 119 |
| Max. P | 0.943023 |

| Location | 22,133,871 – 22,133,990 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -14.26 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
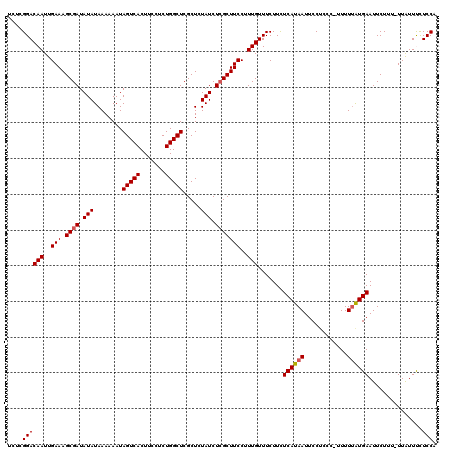
>3L_DroMel_CAF1 22133871 119 + 23771897 UCUCGGACAAUUGAAAGCGAUAUAUAAAAAAUAGUCACUUACUCUGGCUCGCUCUAUCUCGCUUCCUUUGUUUCUUCUCAUAAUUCCUUCC-UUUUUAUGAAUUCUUUUUUGUUUUUCCA ....((((((..(((.((((.(((........(((((.......))))).....))).)))))))..))).......((((((........-...))))))...............))). ( -16.42) >DroSec_CAF1 119730 118 + 1 UCUCGGACAAUUGAAAGCGAUAUAUAAAAAAUAGUCACUUCCUCUGGCUCGCUCUAUCUCGCUUCCUUUGUUUCUUCUCAUAAUUCCUCCC-UUUUUAUGAAUUCUUU-UUAUUCCUCCA ....((((((..(((.((((.(((........(((((.......))))).....))).)))))))..))).......((((((........-...)))))).......-.......))). ( -16.32) >DroSim_CAF1 127800 118 + 1 UCUCGGACAAUUGAAAGCGAUAUAUAAAAAAUAGUCACUUCCUCUGGCUCGCUCUAUCUCGCUUCCUUUGUUUCUUCUCAUAAUUCCUCCC-UUUUUAUGAAUUCUUU-UUAUUCCUCCA ....((((((..(((.((((.(((........(((((.......))))).....))).)))))))..))).......((((((........-...)))))).......-.......))). ( -16.32) >DroEre_CAF1 119550 118 + 1 GCUCGGACAAUUGAAAGCGAUAUAUAAAAAAUAGUCACUUCCUCUGGCUCGCUCUAUCUGGCUUCCCUUGUUUCUUCUCAUAAUUCCUGCCAUUUUGGUGAAUUCUU--UUAUUUCUCCU ....(((.((.((((((.(((............((((.......))))(((((..(..((((.....((((........)))).....))))..).)))))))))))--))).)).))). ( -17.90) >DroYak_CAF1 167160 119 + 1 UCUGGGACAAUUGAAAGCGAUAUAUAAAAAAUAGUCACUUCCUCUGGCUCGCUCUAUCUCGCUUCCUUUGUUUCUUUUCAUAAUUCCGGUCAUUUUUAUGAAUUCUUU-UUAUUUCUCCU ...(..((((..(((.((((.(((........(((((.......))))).....))).)))))))..))))..)..(((((((............)))))))......-........... ( -19.62) >consensus UCUCGGACAAUUGAAAGCGAUAUAUAAAAAAUAGUCACUUCCUCUGGCUCGCUCUAUCUCGCUUCCUUUGUUUCUUCUCAUAAUUCCUCCC_UUUUUAUGAAUUCUUU_UUAUUUCUCCA ....((((((..(((.((((.(((........(((((.......))))).....))).)))))))..))).......((((((............))))))...............))). (-14.26 = -14.50 + 0.24)

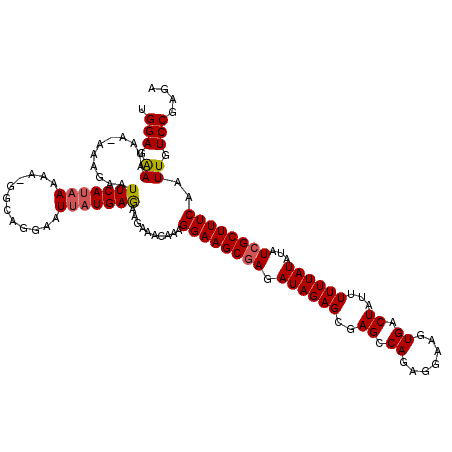
| Location | 22,133,871 – 22,133,990 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -19.54 |
| Energy contribution | -19.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
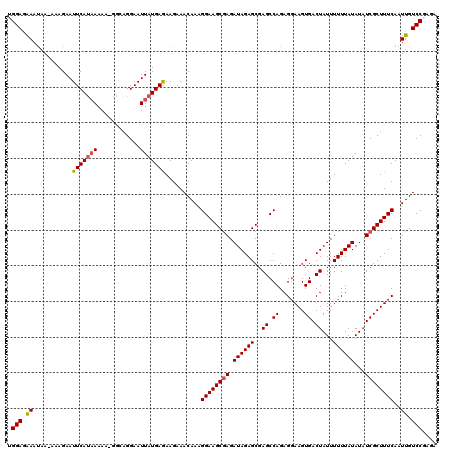
>3L_DroMel_CAF1 22133871 119 - 23771897 UGGAAAAACAAAAAAGAAUUCAUAAAAA-GGAAGGAAUUAUGAGAAGAAACAAAGGAAGCGAGAUAGAGCGAGCCAGAGUAAGUGACUAUUUUUUAUAUAUCGCUUUCAAUUGUCCGAGA .(((..............(((((((...-........)))))))..........((((((((.((((((..((.((.......)).))...))))))...)))))))).....))).... ( -20.40) >DroSec_CAF1 119730 118 - 1 UGGAGGAAUAA-AAAGAAUUCAUAAAAA-GGGAGGAAUUAUGAGAAGAAACAAAGGAAGCGAGAUAGAGCGAGCCAGAGGAAGUGACUAUUUUUUAUAUAUCGCUUUCAAUUGUCCGAGA .(((.((....-......(((((((...-........)))))))..........((((((((.((((((..((.((.......)).))...))))))...))))))))..)).))).... ( -22.20) >DroSim_CAF1 127800 118 - 1 UGGAGGAAUAA-AAAGAAUUCAUAAAAA-GGGAGGAAUUAUGAGAAGAAACAAAGGAAGCGAGAUAGAGCGAGCCAGAGGAAGUGACUAUUUUUUAUAUAUCGCUUUCAAUUGUCCGAGA .(((.((....-......(((((((...-........)))))))..........((((((((.((((((..((.((.......)).))...))))))...))))))))..)).))).... ( -22.20) >DroEre_CAF1 119550 118 - 1 AGGAGAAAUAA--AAGAAUUCACCAAAAUGGCAGGAAUUAUGAGAAGAAACAAGGGAAGCCAGAUAGAGCGAGCCAGAGGAAGUGACUAUUUUUUAUAUAUCGCUUUCAAUUGUCCGAGC .(((.((....--...(((((.((.....))...))))).(((..........((....)).....((((((.....(((((((....))))))).....))))))))).)).))).... ( -21.90) >DroYak_CAF1 167160 119 - 1 AGGAGAAAUAA-AAAGAAUUCAUAAAAAUGACCGGAAUUAUGAAAAGAAACAAAGGAAGCGAGAUAGAGCGAGCCAGAGGAAGUGACUAUUUUUUAUAUAUCGCUUUCAAUUGUCCCAGA .(((.((....-......(((((((............)))))))..........((((((((.((((((..((.((.......)).))...))))))...))))))))..)).))).... ( -21.30) >consensus UGGAGAAAUAA_AAAGAAUUCAUAAAAA_GGCAGGAAUUAUGAGAAGAAACAAAGGAAGCGAGAUAGAGCGAGCCAGAGGAAGUGACUAUUUUUUAUAUAUCGCUUUCAAUUGUCCGAGA .(((.((...........(((((((............)))))))..........((((((((.((((((..((.((.......)).))...))))))...))))))))..)).))).... (-19.54 = -19.74 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:07 2006