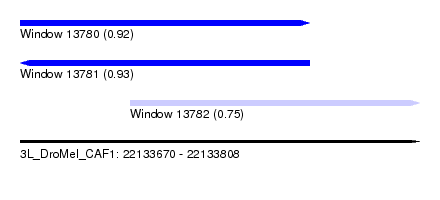
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,133,670 – 22,133,808 |
| Length | 138 |
| Max. P | 0.929580 |

| Location | 22,133,670 – 22,133,770 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -17.93 |
| Consensus MFE | -11.47 |
| Energy contribution | -12.05 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22133670 100 + 23771897 AAUUUUUCGUCUU--UAUAAUUUAAUUUUGCUAUAAAAACGCAUUAUUGUAUAAUGCGUUGGGCUAUCUCUGACGUCCCCGCCC--CCUUACCUACAUCAAAUG .......((((.(--((.....)))....(((.....((((((((((...)))))))))).))).......)))).........--.................. ( -15.00) >DroGri_CAF1 136987 81 + 1 AAUUUUUCGUCUUUUUAUAAUUUCAUUUUACUAUGAAAACGCAUUAUUAUGUAAUGCGCUGGGCUG-CGCU--CCGCGACGCUC-------------------- .......((((.........((((((......))))))..(((((((...)))))))((.((((..-.)))--).))))))...-------------------- ( -19.70) >DroSec_CAF1 119492 100 + 1 AAUUUUUCGUCUU--UAUAAUUUAAUUUUGCUAUAAAAACGCAUUAUUGUAUAAUGCGUUGGGCUAUCUCUGACUGCCCCGCCC--CCUUACCAACAUCAAACG .............--..............((......((((((((((...))))))))))((((..((...))..)))).))..--.................. ( -17.40) >DroSim_CAF1 127568 100 + 1 AAUUUUUCGUCUU--UAUAAUUUAAUUUUGCUAUAAAAACGCAUUAUGGUAUAAUGCGUUGGGCUAUGUCUGACUGCCCCGCCC--CCUUACCUACAUCAAACG .............--..............((......((((((((((...))))))))))((((...........)))).))..--.................. ( -17.00) >DroEre_CAF1 119367 88 + 1 AAUUUUUCGUCUU--UAUAAUUUCAUUUUGCUAUAACAACGCAUUAUUGUAUAAUGCGUUGGGCU-AUUCUCACAGCCCC-------------ACCCCCCAGCG .............--..............(((....(((((((((((...)))))))))))((((-........))))..-------------.......))). ( -20.50) >DroYak_CAF1 166958 101 + 1 AAUUUUUCGUCUU--UAUAAUUUAAUUUUGCUAUAAAAACGCAUUAUUGUAUAAUGCGUUGGGCUUGCUCUCUCAGCUACGCCAAACGCUA-CUCCCCCAAACG .............--..............((......((((((((((...)))))))))).(((..(((.....)))...)))....))..-............ ( -18.00) >consensus AAUUUUUCGUCUU__UAUAAUUUAAUUUUGCUAUAAAAACGCAUUAUUGUAUAAUGCGUUGGGCUAUCUCUGACAGCCCCGCCC__CCUUA_CUACAUCAAACG .....................................((((((((((...))))))))))((((...........))))......................... (-11.47 = -12.05 + 0.58)


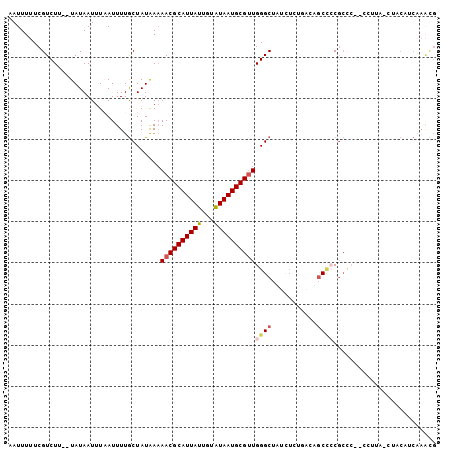
| Location | 22,133,670 – 22,133,770 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -11.80 |
| Energy contribution | -12.72 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22133670 100 - 23771897 CAUUUGAUGUAGGUAAGG--GGGCGGGGACGUCAGAGAUAGCCCAACGCAUUAUACAAUAAUGCGUUUUUAUAGCAAAAUUAAAUUAUA--AAGACGAAAAAUU ..((((.((((((...((--(((((....))))........)))((((((((((...)))))))))))))))).))))...........--............. ( -23.00) >DroGri_CAF1 136987 81 - 1 --------------------GAGCGUCGCGG--AGCG-CAGCCCAGCGCAUUACAUAAUAAUGCGUUUUCAUAGUAAAAUGAAAUUAUAAAAAGACGAAAAAUU --------------------...((((..((--.((.-..)))).((((((((.....))))))))((((((......)))))).........))))....... ( -18.60) >DroSec_CAF1 119492 100 - 1 CGUUUGAUGUUGGUAAGG--GGGCGGGGCAGUCAGAGAUAGCCCAACGCAUUAUACAAUAAUGCGUUUUUAUAGCAAAAUUAAAUUAUA--AAGACGAAAAAUU (((((........(((..--..((.((((.(((...))).))))((((((((((...))))))))))......))....))).......--.)))))....... ( -25.29) >DroSim_CAF1 127568 100 - 1 CGUUUGAUGUAGGUAAGG--GGGCGGGGCAGUCAGACAUAGCCCAACGCAUUAUACCAUAAUGCGUUUUUAUAGCAAAAUUAAAUUAUA--AAGACGAAAAAUU (((((..(((((.(((..--..((.((((...........))))((((((((((...))))))))))......))....)))..)))))--.)))))....... ( -24.00) >DroEre_CAF1 119367 88 - 1 CGCUGGGGGGU-------------GGGGCUGUGAGAAU-AGCCCAACGCAUUAUACAAUAAUGCGUUGUUAUAGCAAAAUGAAAUUAUA--AAGACGAAAAAUU .((((......-------------.(((((((....))-)))))((((((((((...))))))))))....))))..............--............. ( -26.20) >DroYak_CAF1 166958 101 - 1 CGUUUGGGGGAG-UAGCGUUUGGCGUAGCUGAGAGAGCAAGCCCAACGCAUUAUACAAUAAUGCGUUUUUAUAGCAAAAUUAAAUUAUA--AAGACGAAAAAUU ((((..(...((-(.(((((.(((...(((.....)))..))).))))))))...)...))))(((((((((((..........)))))--))))))....... ( -24.10) >consensus CGUUUGAUGUAG_UAAGG__GGGCGGGGCAGUCAGAGAUAGCCCAACGCAUUAUACAAUAAUGCGUUUUUAUAGCAAAAUUAAAUUAUA__AAGACGAAAAAUU ......................((.((((...........))))(((((((((.....)))))))))......))............................. (-11.80 = -12.72 + 0.92)

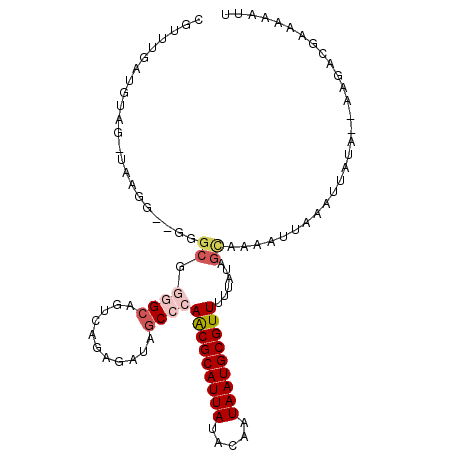
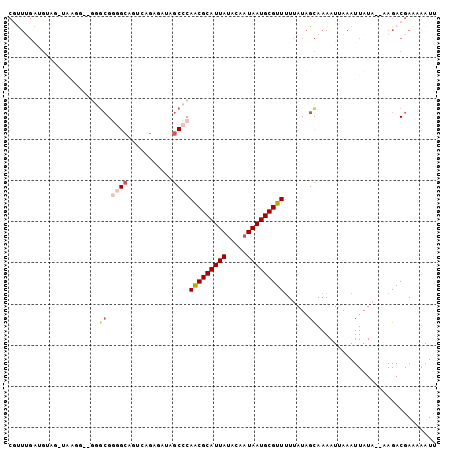
| Location | 22,133,708 – 22,133,808 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -24.74 |
| Consensus MFE | -17.44 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22133708 100 + 23771897 GCAUUAUUGUAUAAUGCGUUGGGCUAUCUCUGACGUCCCCGCCC--CCUUACCUACAUCAAAUGCAGAGUACCAUCAUCAGCGUCAGCAUUCGCAUC-AGCAU- (((((((...)))))))(((((((.....(((((((........--...(((((.(((...))).)).))).........))))))).....)).))-)))..- ( -22.35) >DroSec_CAF1 119530 101 + 1 GCAUUAUUGUAUAAUGCGUUGGGCUAUCUCUGACUGCCCCGCCC--CCUUACCAACAUCAAACGCAGAGUGCUAUCAUCAGCGUCAGCAUUCGCAUCCAGCAU- (((((((...)))))))((.((((..((...))..)))).))..--.................((.(((((((..(......)..))))))))).........- ( -24.70) >DroSim_CAF1 127606 102 + 1 GCAUUAUGGUAUAAUGCGUUGGGCUAUGUCUGACUGCCCCGCCC--CCUUACCUACAUCAAACGCAGAGUGCCAUCAUCAGCGUCAGCAUUCGCAUCCAGCAUC ((...(((((((..((((((((((...((......))...))))--..............))))))..))))))).....(((........))).....))... ( -26.03) >DroEre_CAF1 119405 82 + 1 GCAUUAUUGUAUAAUGCGUUGGGCU-AUUCUCACAGCCCC-------------ACCCCCCAGCGCAGAGUGCCAUCAUCAGCGACAGCACUCGCAU-------- (((((((...)))))))((.(((((-........))))).-------------))........((.((((((..((......))..))))))))..-------- ( -27.40) >DroYak_CAF1 166996 95 + 1 GCAUUAUUGUAUAAUGCGUUGGGCUUGCUCUCUCAGCUACGCCAAACGCUA-CUCCCCCAAACGCAGAGUGCCAUCAUCAGCGUCAGCAUUCGCAU-------- (((((((...)))))))(((.(((..(((.....)))...))).)))....-...........((.((((((...(......)...))))))))..-------- ( -23.20) >consensus GCAUUAUUGUAUAAUGCGUUGGGCUAUCUCUGACAGCCCCGCCC__CCUUACCUACAUCAAACGCAGAGUGCCAUCAUCAGCGUCAGCAUUCGCAUC_AGCAU_ (((((((...)))))))((.((((...........)))).)).....................((.((((((...(......)...)))))))).......... (-17.44 = -18.12 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:05 2006