
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,130,316 – 22,130,462 |
| Length | 146 |
| Max. P | 0.806330 |

| Location | 22,130,316 – 22,130,426 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -19.08 |
| Energy contribution | -18.67 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
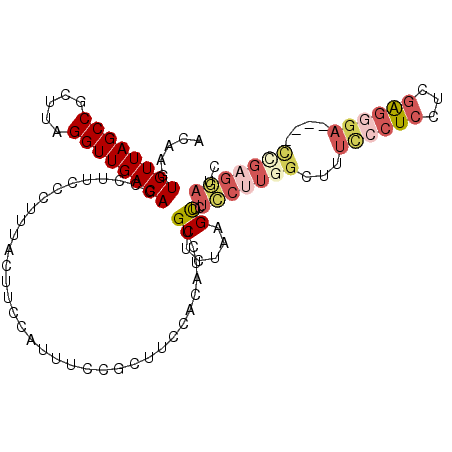
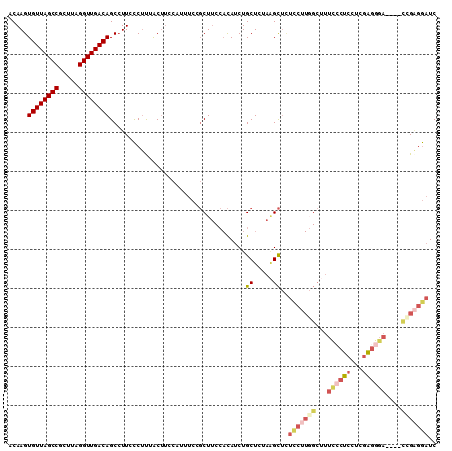
>3L_DroMel_CAF1 22130316 110 + 23771897 CCGCUUGUGUUUGUGUUUUCAUCAAAUAAAAUUGACACACACAAGUGUUAGCCGCUUAGGUUGACAGCCUUCCCUUU--------AC-UUCCAUUUCCGCUU-CCACAUCUGCUCUAAGC ..((((((((..(((((................)))))))))))))(((((((.....)))))))............--------..-..........((..-........))....... ( -22.49) >DroGri_CAF1 133868 118 + 1 CUGUUUGUGUUUGUAUUUUCUUCAAAUAAAAUUGACACACACAAGUGUUAGCCGCUUUAGCUGACAGUAUUUGCAAAAAAAAAAAACUUUCGAGGACUGCUGGCAACAUUUGUGAGAC-- .....((((((...(((((........))))).))))))((((((((((.(((((....))...((((.((((.(((..........))))))).))))..)))))))))))))....-- ( -28.10) >DroSec_CAF1 116102 110 + 1 CCGCUUGUGUUUGUGUUUUCAUCAAAUAAAAUUGACACACACAAGUGUUAGCCGCUUAGGUUGACAGCCUUCCCUUU--------AC-UUCCAUUUCCGCUG-CCACAUCUGCUCUAAGC .(((((((((..(((((................))))))))))))))......((((((((.((((((.........--------..-..........))))-.....)).)).)))))) ( -24.79) >DroEre_CAF1 114989 110 + 1 CCGCUUGUGUUUGUGUUUUCAUCAAAUAAAAUUGACACACACAAGUGUUAGCCGCUUAGGUUGACAGCCUUUCCUCU--------GC-UUCCAUUUCCGCUU-CCACAUCUGCUAUAAGC ..((((((((..(((((................)))))))))))))(((((((.....)))))))(((.........--------((-..........))..-........)))...... ( -23.40) >DroYak_CAF1 163721 110 + 1 CUGCUUGUGUUUGUGUUUUCAUCAAAUAAAAUUGACACACACAAGUGUUAGCCGCUUAGGUUGACAGCCUUAUCCUU--------AC-UUCCAUUUCCGCUU-CCACAUCUGCUAUAAGC ..((((((((..(((((................)))))))))))))(((((((.....)))))))............--------..-..........((..-........))....... ( -22.39) >DroAna_CAF1 133438 100 + 1 CUGUUUGUGUUUGUAUUUUCAUCAAAUAAAAUUGACACACACAAGUGUUAGCCGCUUAGGUUGACAGC-UUCCCUCC--------AC-A------UCCUCUU----UAUCUGCUCGAGGU .(((.((((((...(((((........))))).)))))).)))..((((((((.....))))))))..-........--------..-.------.((((..----.........)))). ( -21.80) >consensus CCGCUUGUGUUUGUGUUUUCAUCAAAUAAAAUUGACACACACAAGUGUUAGCCGCUUAGGUUGACAGCCUUCCCUUU________AC_UUCCAUUUCCGCUU_CCACAUCUGCUAUAAGC ..((((((((.((((((((........)))))..))).))))))))(((((((.....)))))))....................................................... (-19.08 = -18.67 + -0.42)



| Location | 22,130,356 – 22,130,462 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -18.25 |
| Energy contribution | -19.87 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22130356 106 + 23771897 ACAAGUGUUAGCCGCUUAGGUUGACAGCCUUCCCUUUACUUCCAUUUCCGCUUCCACAUCUGCUCUAAGCUUUCCUUGGCUUUCCCUCCUCGAGGGA----CCGAGGAUC .....((((((((.....)))))))).......................((((.............))))..(((((((...((((((...))))))----))))))).. ( -29.42) >DroSec_CAF1 116142 106 + 1 ACAAGUGUUAGCCGCUUAGGUUGACAGCCUUCCCUUUACUUCCAUUUCCGCUGCCACAUCUGCUCUAAGCUCUCCUUGGCUUUCCCUCGUCGGGGGA----UCGAGGAUC .....((((((((.....))))))))......((((....(((.........((((.....((.....))......))))...(((.....))))))----..))))... ( -27.50) >DroSim_CAF1 124265 106 + 1 ACAAGUGUUAGCCGCUUAGGUUGACAGCCUUCCCUUUACUUCCAUUUCCGCUUCCACAUCUGCUCUAUGCUCUCCUUGGCUUUCCCUCGUCGGGGGA----CCGAGGAUC .....((((((((.....))))))))...................................((.....))..(((((((.(..(((.....)))..)----))))))).. ( -29.40) >DroEre_CAF1 115029 105 + 1 ACAAGUGUUAGCCGCUUAGGUUGACAGCCUUUCCUCUGCUUCCAUUUCCGCUUCCACAUCUGCUAUAAGCUCUCCUUCGCUUUU-CUCCUCCAAGGA----CCGAGGAUC .....((((((((.....)))))))).....(((((.(..(((......((..........))...((((........))))..-.........)))----).))))).. ( -22.30) >DroYak_CAF1 163761 105 + 1 ACAAGUGUUAGCCGCUUAGGUUGACAGCCUUAUCCUUACUUCCAUUUCCGCUUCCACAUCUGCUAUAAGCUCUUCUUGGCUUUC-CUCCUCGAGGGA----CUGUGAAUC .....((((((((.....))))))))............................((((...((((...........)))).(((-(((...))))))----.)))).... ( -20.80) >DroAna_CAF1 133478 87 + 1 ACAAGUGUUAGCCGCUUAGGUUGACAGC-UUCCCUCCACA------UCCUCUU---UAUCUGCUCGAGGU-------------CCCUCGAGGAGGCAAGCCACGGAGCUC ..(((((((((((.....)))))))).)-))..((((...------.......---......((((((..-------------..))))))..((....))..))))... ( -29.00) >consensus ACAAGUGUUAGCCGCUUAGGUUGACAGCCUUCCCUUUACUUCCAUUUCCGCUUCCACAUCUGCUCUAAGCUCUCCUUGGCUUUCCCUCCUCGAGGGA____CCGAGGAUC .....((((((((.....))))))))...................................((.....))..(((((((...((((((...))))))....))))))).. (-18.25 = -19.87 + 1.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:15:00 2006