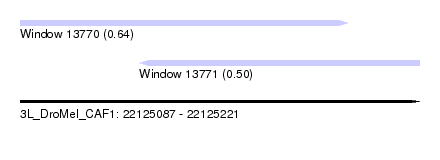
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,125,087 – 22,125,221 |
| Length | 134 |
| Max. P | 0.635194 |

| Location | 22,125,087 – 22,125,197 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.53 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22125087 110 + 23771897 CUCCCCCUCCUCAACCGAUAAACCCAGCCCAACCAGACCUGGCCGCAAUUUUCGUUGCGUUUGCAAAAUAUUUCCAAGUGUCUUGGCCGCUCAAUUUAUGAGCGUUGAAA ..........(((((.(.(((((((((..(.....)..))))..(((((....))))))))))).........(((((...)))))..(((((.....)))))))))).. ( -23.70) >DroSec_CAF1 110886 94 + 1 CUCUCCCUC----------------AUCCCAACCAGGCCUGGCCGCAAUUUUCGUUGCGUUUGCAAAAUAUUCGCUGGUGUCUCACCAGCUCAAUUUAUGAGCGUUGAAA .((..((((----------------((..(((.(.(((...)))(((((....)))))).)))..........(((((((...))))))).......))))).)..)).. ( -23.30) >DroSim_CAF1 118977 94 + 1 CUCUCCCUC----------------AUCCCAACCAGGCCUGGCCGCAAUUUUCGUUGCGUUUGCAAAAUAUUCGCUGGUGUCUCACCAGCUCAAUUUAUGAGCGUUGAAA .((..((((----------------((..(((.(.(((...)))(((((....)))))).)))..........(((((((...))))))).......))))).)..)).. ( -23.30) >DroEre_CAF1 109804 105 + 1 CUCCCCC-----AAUCAACAAAGCCAGCCCAACCAGGCCAAGCCGCAAUUUUCAUUGCGUUUGCAAAAUAUUUGCUGGUGUCUUGCCGGCUCAAUUUAUGAGCGUUGAAA .......-----..(((((...((((((........((.((((.(((((....))))))))))).........)))))).........(((((.....)))))))))).. ( -29.73) >DroYak_CAF1 153569 104 + 1 CUCCCCC-----AAUCGCCA-ACCCAGCCCAACCAGGCCAGGCCGCAAUUUUCGUUGCGUUUGCAAGAUAUUUGCCGGUGUCUUAGCCGCUCAAUUUAUGAGCGUUGAAA .......-----......((-((...(((......)))..(((((((((....)))))).....(((((((......))))))).)))(((((.....)))))))))... ( -28.20) >DroAna_CAF1 128593 105 + 1 CACCGCC-----AAUCAUUUUCCCUCGCAUGCCGCGGCCUGGCCGCAAUUUUCAUUGCGUUUGCAAAAUAUUUUCUGGUGUCUUGGCCCCGCGAUUUAUGAGCGUUGAAA ((.(((.-----..((((......((((..((((((((...)))))........((((....))))..................)))...))))...))))))).))... ( -26.20) >consensus CUCCCCC_____AAUC___A_ACCCAGCCCAACCAGGCCUGGCCGCAAUUUUCGUUGCGUUUGCAAAAUAUUUGCUGGUGUCUUACCCGCUCAAUUUAUGAGCGUUGAAA .............................((((..((((((((((((((....))))))..............))))).)))......(((((.....)))))))))... (-16.28 = -16.53 + 0.25)

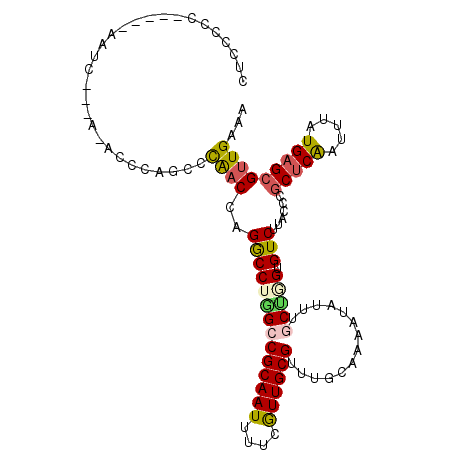
| Location | 22,125,127 – 22,125,221 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.38 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
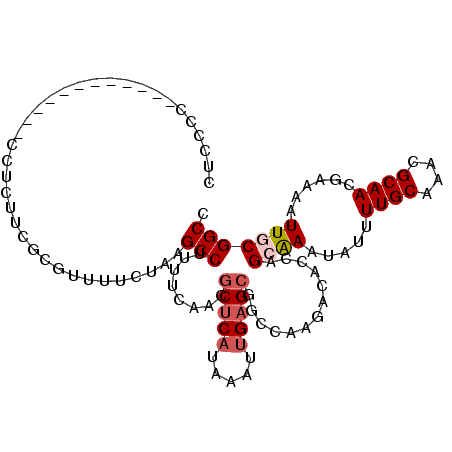
>3L_DroMel_CAF1 22125127 94 - 23771897 CUCGCC----------------UUCGCGUUUUCUAAGCCUUUUCAACGCUCAUAAAUUGAGCGGCCAAGACACUUGGAAAUAUUUUGCAAACGCAACGAAAAUUGCGGCC ...(((----------------...(((.((((((((..((((...((((((.....))))))...))))..)))))))).....)))....((((......))))))). ( -28.40) >DroSec_CAF1 110910 98 - 1 CUCCCC------------CCUUUUCGCGUUUUCUAAGCCUUUUCAACGCUCAUAAAUUGAGCUGGUGAGACACCAGCGAAUAUUUUGCAAACGCAACGAAAAUUGCGGCC ......------------.....((((((((((...((.........(((((.....)))))(((((...))))))).......((((....)))).))))).))))).. ( -24.10) >DroSim_CAF1 119001 98 - 1 CUCCCC------------CCUUUUCGCGUUUUCUAAGCCUUUUCAACGCUCAUAAAUUGAGCUGGUGAGACACCAGCGAAUAUUUUGCAAACGCAACGAAAAUUGCGGCC ......------------.....((((((((((...((.........(((((.....)))))(((((...))))))).......((((....)))).))))).))))).. ( -24.10) >DroEre_CAF1 109839 105 - 1 CUCCCCCCACCGC-----CCUCUUCGCGAUUUCUAAGCCUUUUCAACGCUCAUAAAUUGAGCCGGCAAGACACCAGCAAAUAUUUUGCAAACGCAAUGAAAAUUGCGGCU ...........((-----(......((.........)).........(((((.....))))).))).........((((.....))))...((((((....))))))... ( -24.10) >DroYak_CAF1 153603 94 - 1 CUCC----------------ACUUCCCGAUUUCUAAGCCUUUUCAACGCUCAUAAAUUGAGCGGCUAAGACACCGGCAAAUAUCUUGCAAACGCAACGAAAAUUGCGGCC ..((----------------......((...(((.((((........(((((.....))))))))).)))...))((((.(.((((((....)))).)).).)))))).. ( -22.60) >DroAna_CAF1 128628 107 - 1 CUCCCA---CCUCCCAUCCCCCACCACUCGUUCUAAGCCUUUUCAACGCUCAUAAAUCGCGGGGCCAAGACACCAGAAAAUAUUUUGCAAACGCAAUGAAAAUUGCGGCC ......---.................((.(((((..((((......(((.........)))))))..))).)).))...............((((((....))))))... ( -18.20) >consensus CUCCCC____________CCUCUUCGCGUUUUCUAAGCCUUUUCAACGCUCAUAAAUUGAGCGGCCAAGACACCAGCAAAUAUUUUGCAAACGCAACGAAAAUUGCGGCC ....................................(((........(((((.....))))).............((((.....((((....))))......))))))). (-13.31 = -13.92 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:55 2006