
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,124,384 – 22,124,552 |
| Length | 168 |
| Max. P | 0.992965 |

| Location | 22,124,384 – 22,124,475 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -19.02 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22124384 91 - 23771897 GCC-------AUCUACGCAAUAUUCUGCAAUCAAGUGUGCAUAUUUACUUCCUGUAAUUGAGCAUUUAUGGCUUUUAUGGCCUAACGGAUUUGCCCGA (((-------((....(((......)))..(((...((((.((.((((.....)))).)).))))...))).....)))))....(((......))). ( -19.50) >DroSec_CAF1 110190 91 - 1 GCC-------AUCUACGCAAUAUUCUGCAAUCAAGUGUGCAUAUUUACUUCCUGUAAUUGAGCAUUUAUGGCUUUUAUGGCCUAACGGAUUUGCCCGU (((-------((....(((......)))..(((...((((.((.((((.....)))).)).))))...))).....)))))...((((......)))) ( -19.90) >DroSim_CAF1 118283 91 - 1 GCC-------AUCUACGCAAUAUUCUGCAAUCAAGUGUGCAUAUUUACUUCCUGUAAUUGAGCAUUUAUGGCUUUUAUGGCCUAACGGAUUUGCCCGU (((-------((....(((......)))..(((...((((.((.((((.....)))).)).))))...))).....)))))...((((......)))) ( -19.90) >DroEre_CAF1 109101 91 - 1 GCC-------AUCUACGCAAUAUUCUGCAAUCAAGUGUGCAUAUUUACUUCCUGUAAUUGAGCAUUUAUGGCUUUUAUGGCCUAACGGAUUUGCCCGU (((-------((....(((......)))..(((...((((.((.((((.....)))).)).))))...))).....)))))...((((......)))) ( -19.90) >DroYak_CAF1 152868 91 - 1 GCC-------AUCUACGCAAUAUUCUGCAAUCAAGUGUGCAUAUUUACUUCCUGUAAUUGAGCAUUUAUGGCUUUUAUGGCCUAACGGAUUUGCCCGU (((-------((....(((......)))..(((...((((.((.((((.....)))).)).))))...))).....)))))...((((......)))) ( -19.90) >DroAna_CAF1 127937 98 - 1 GCCAGCCGUAAUCUACGCAACAUUCUGCAAUCAAGUGUGCAUAUUUACUUCCUGUAAUUGAGCAUUUACGGCUUUUAUGGCCUAACGGAUUUGUCCCC (((((((((((.....(((......)))........((((.((.((((.....)))).)).))))))))))).....)))).....(((....))).. ( -24.90) >consensus GCC_______AUCUACGCAAUAUUCUGCAAUCAAGUGUGCAUAUUUACUUCCUGUAAUUGAGCAUUUAUGGCUUUUAUGGCCUAACGGAUUUGCCCGU ................((((...((((.....((((((.((...((((.....)))).)).))))))..((((.....))))...)))).)))).... (-19.02 = -18.88 + -0.14)

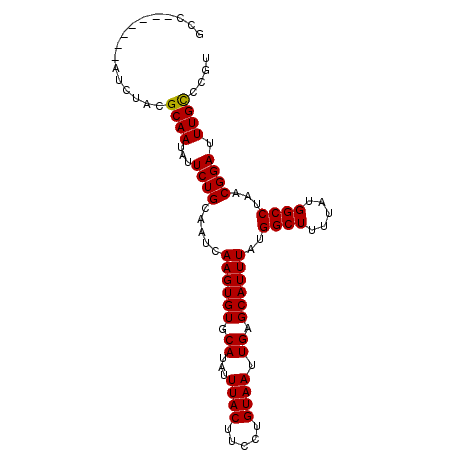

| Location | 22,124,442 – 22,124,552 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.82 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.62 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22124442 110 + 23771897 GCACACUUGAUUGCAGAAUAUUGCGUAGAUGGCCAGGCCCUUGGUUCUGGCUUUUACUCAGUCUCUGGCUAAGACUCUGGCCUAAGCUAAGACUUGUCCUGGCCAAGUUG ....(((((....((((..((((.(((((.((((((..(....)..))))))))))).)))).))))((((.(((((((((....))).)))...))).))))))))).. ( -36.20) >DroSec_CAF1 110248 110 + 1 GCACACUUGAUUGCAGAAUAUUGCGUAGAUGGCCAGGCCUUUGGUUCUGGCUUUUACUCAGUCUUUGGCUAAGACUCCGGUCUGAGCUGAGACUUGUCCUGGCCAAGUUG .....((....(((((....))))).)).((((((((.(...((((..(((((..(((.((((((.....))))))..)))..)))))..)))).).))))))))..... ( -37.20) >DroSim_CAF1 118341 110 + 1 GCACACUUGAUUGCAGAAUAUUGCGUAGAUGGCCAGGCCUUUGGUUCUGGCUUUUACUCAGUCUUUGGCUAAGACUCCGGUCUGAGCUGAGACUUGUCCUGGCCAAGUUG .....((....(((((....))))).)).((((((((.(...((((..(((((..(((.((((((.....))))))..)))..)))))..)))).).))))))))..... ( -37.20) >DroEre_CAF1 109159 110 + 1 GCACACUUGAUUGCAGAAUAUUGCGUAGAUGGCCAGGCCUUUGGUUCUGGCUUUUACGCAGUCUCGGGCUAAGACUGCGGCCUCAACUGAGACUUGUCCUGGCCAAGUUG (((........))).((..((((((((((.(((((((((...)).))))))))))))))))).)).(((((.(((...((.(((....))).)).))).)))))...... ( -40.00) >DroYak_CAF1 152926 110 + 1 GCACACUUGAUUGCAGAAUAUUGCGUAGAUGGCCAGGCCUUGGGUUCUGGCUUUUACUCAGUCUCUGGCUCAGACUGCAGCUUUCGCAGAGACUUGUCCUGGUCAAGUUG ....(((((((..(((.(((.((.(((((.(((((((((...)).)))))))))))).))(((((((((((.....).))).....))))))).))).)))))))))).. ( -35.50) >consensus GCACACUUGAUUGCAGAAUAUUGCGUAGAUGGCCAGGCCUUUGGUUCUGGCUUUUACUCAGUCUCUGGCUAAGACUCCGGCCUGAGCUGAGACUUGUCCUGGCCAAGUUG ....(((((....((((..((((.(((((.((((((..(....)..))))))))))).)))).))))((((.(((..((((....))))......))).))))))))).. (-30.70 = -30.62 + -0.08)


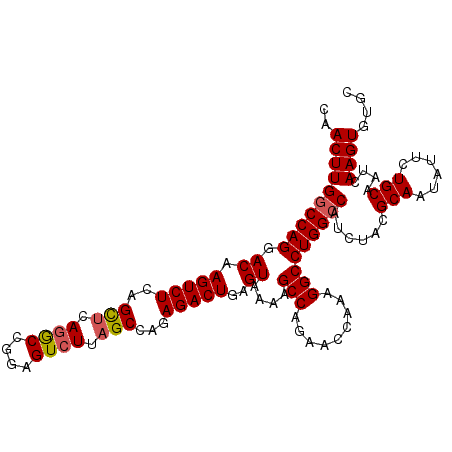
| Location | 22,124,442 – 22,124,552 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.82 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -29.26 |
| Energy contribution | -29.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22124442 110 - 23771897 CAACUUGGCCAGGACAAGUCUUAGCUUAGGCCAGAGUCUUAGCCAGAGACUGAGUAAAAGCCAGAACCAAGGGCCUGGCCAUCUACGCAAUAUUCUGCAAUCAAGUGUGC ..(((((((((((((.((((((.(((.((((....)))).)))..))))))..)).....((........)).)))))))......(((......)))....)))).... ( -34.20) >DroSec_CAF1 110248 110 - 1 CAACUUGGCCAGGACAAGUCUCAGCUCAGACCGGAGUCUUAGCCAAAGACUGAGUAAAAGCCAGAACCAAAGGCCUGGCCAUCUACGCAAUAUUCUGCAAUCAAGUGUGC ..((((((((((.((.(((((..(((.((((....)))).)))...)))))..))....(((.........)))))))))......(((......)))....)))).... ( -32.60) >DroSim_CAF1 118341 110 - 1 CAACUUGGCCAGGACAAGUCUCAGCUCAGACCGGAGUCUUAGCCAAAGACUGAGUAAAAGCCAGAACCAAAGGCCUGGCCAUCUACGCAAUAUUCUGCAAUCAAGUGUGC ..((((((((((.((.(((((..(((.((((....)))).)))...)))))..))....(((.........)))))))))......(((......)))....)))).... ( -32.60) >DroEre_CAF1 109159 110 - 1 CAACUUGGCCAGGACAAGUCUCAGUUGAGGCCGCAGUCUUAGCCCGAGACUGCGUAAAAGCCAGAACCAAAGGCCUGGCCAUCUACGCAAUAUUCUGCAAUCAAGUGUGC ..((((((.(((((..((((((.((((((((....))))))))..))))))(((((...(((((..(....)..)))))....)))))....)))))...)))))).... ( -40.20) >DroYak_CAF1 152926 110 - 1 CAACUUGACCAGGACAAGUCUCUGCGAAAGCUGCAGUCUGAGCCAGAGACUGAGUAAAAGCCAGAACCCAAGGCCUGGCCAUCUACGCAAUAUUCUGCAAUCAAGUGUGC ..((((((.(((((..(((((((((....)).((.......)).)))))))..(((...(((((..(....)..)))))....)))......)))))...)))))).... ( -32.30) >consensus CAACUUGGCCAGGACAAGUCUCAGCUCAGGCCGGAGUCUUAGCCAGAGACUGAGUAAAAGCCAGAACCAAAGGCCUGGCCAUCUACGCAAUAUUCUGCAAUCAAGUGUGC ..((((((((((.((.(((((..(((.((((....)))).)))...)))))..))....(((.........)))))))))......(((......)))....)))).... (-29.26 = -29.46 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:52 2006