
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,123,491 – 22,123,600 |
| Length | 109 |
| Max. P | 0.998531 |

| Location | 22,123,491 – 22,123,600 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.52 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -4.70 |
| Energy contribution | -5.58 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.24 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22123491 109 + 23771897 CCCCUCUGGUUCUGGUUU-UGGUUCUCCUCCAGCAAUUUCUCUGCUCUCGCAUCGAAAACACC-----AAACCAAAUAAAUAAUAGUUAAAUUUU-UUCUAUUAUAUCUUUCCAGA ....(((((...((((((-(((.......)))....((((..(((....)))..)))).....-----)))))).....(((((((.........-..)))))))......))))) ( -19.70) >DroGri_CAF1 124680 106 + 1 CCAUUCCCAUUCCCACUUCCAAAUCCCUCCCACAAAUUUCAGUGACUACGCAUUGAGUGCCC--------AGCGAAUAAAUAAUAGUUAAAUUUU--UCGUUCAUUUCCUAUAGAA .....................................(((((((......))))))).....--------((((((.((((.........)))))--))))).............. ( -8.20) >DroSec_CAF1 109589 113 + 1 C-CCUCUGGUUCUGGUUU-CGGUUCUCCUCCAGCAAUUUCUGUGCUCGCGCAUCGAAAGCACCAACCCAAACCAAAUAAAUAAUAGUUAAAUUUU-UUCUAUUAUAUCUUUCCAGA .-..(((((...((((((-.((((........((..((((.((((....)))).))))))...)))).)))))).....(((((((.........-..)))))))......))))) ( -24.00) >DroSim_CAF1 117369 114 + 1 CCCCUCUGGUUCUGGUUU-CGGUUCUCCUCCAGCAAUUUCUCUGCUCGCGCAUCGAAAGCACCAACCCAAACCAAAUAAAUAAUAGUUAAAUUUU-UUCUAUUAUAUCUUUCCAGA ....(((((...((((((-.((((.......((((.......)))).((.........))...)))).)))))).....(((((((.........-..)))))))......))))) ( -22.50) >DroYak_CAF1 151967 99 + 1 CCCCUCUGGUUCUAGUUU-UGGUUGUCCUCCAGCCAGUUCUCUGCUCGCA-----------CC-----CAACCAAAUAAAUAAUAGUUAAAUUUCUUUCUAUUAUUGCUUUCCAGA ....(((((....(((((-((((((....(.(((.((....))))).)..-----------..-----))))))))..((((((((............)))))))))))..))))) ( -22.10) >consensus CCCCUCUGGUUCUGGUUU_CGGUUCUCCUCCAGCAAUUUCUCUGCUCGCGCAUCGAAAGCACC_____AAACCAAAUAAAUAAUAGUUAAAUUUU_UUCUAUUAUAUCUUUCCAGA ....(((((...(((((...((....))..(((........))).........................))))).....(((((((............)))))))......))))) ( -4.70 = -5.58 + 0.88)



| Location | 22,123,491 – 22,123,600 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.52 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.74 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22123491 109 - 23771897 UCUGGAAAGAUAUAAUAGAA-AAAAUUUAACUAUUAUUUAUUUGGUUU-----GGUGUUUUCGAUGCGAGAGCAGAGAAAUUGCUGGAGGAGAACCA-AAACCAGAACCAGAGGGG (((((......(((((((..-.........)))))))...((((((((-----(((..((((..(((....)))..))))..)))...((....)).-)))))))).))))).... ( -29.40) >DroGri_CAF1 124680 106 - 1 UUCUAUAGGAAAUGAACGA--AAAAUUUAACUAUUAUUUAUUCGCU--------GGGCACUCAAUGCGUAGUCACUGAAAUUUGUGGGAGGGAUUUGGAAGUGGGAAUGGGAAUGG ...................--.........(((((.((((((((((--------(.(((.....))).))))((((.((((((.......))))))...)))).)))))))))))) ( -17.00) >DroSec_CAF1 109589 113 - 1 UCUGGAAAGAUAUAAUAGAA-AAAAUUUAACUAUUAUUUAUUUGGUUUGGGUUGGUGCUUUCGAUGCGCGAGCACAGAAAUUGCUGGAGGAGAACCG-AAACCAGAACCAGAGG-G (((((......(((((((..-.........)))))))...((((((((.(((((((..((((..(((....)))..))))..))).......)))).-)))))))).)))))..-. ( -33.71) >DroSim_CAF1 117369 114 - 1 UCUGGAAAGAUAUAAUAGAA-AAAAUUUAACUAUUAUUUAUUUGGUUUGGGUUGGUGCUUUCGAUGCGCGAGCAGAGAAAUUGCUGGAGGAGAACCG-AAACCAGAACCAGAGGGG (((((......(((((((..-.........)))))))...((((((((.(((((((..((((..(((....)))..))))..))).......)))).-)))))))).))))).... ( -34.71) >DroYak_CAF1 151967 99 - 1 UCUGGAAAGCAAUAAUAGAAAGAAAUUUAACUAUUAUUUAUUUGGUUG-----GG-----------UGCGAGCAGAGAACUGGCUGGAGGACAACCA-AAACUAGAACCAGAGGGG (((((.....(((((((((((....)))..))))))))..((((((((-----..-----------..(.(((((....)).))).)....))))))-)).......))))).... ( -27.10) >consensus UCUGGAAAGAUAUAAUAGAA_AAAAUUUAACUAUUAUUUAUUUGGUUU_____GGUGCUUUCGAUGCGCGAGCAGAGAAAUUGCUGGAGGAGAACCG_AAACCAGAACCAGAGGGG (((((......(((((((............)))))))...(((((((......((((.((((..(((....)))..)))).))))...((....))...))))))).))))).... (-14.86 = -15.74 + 0.88)

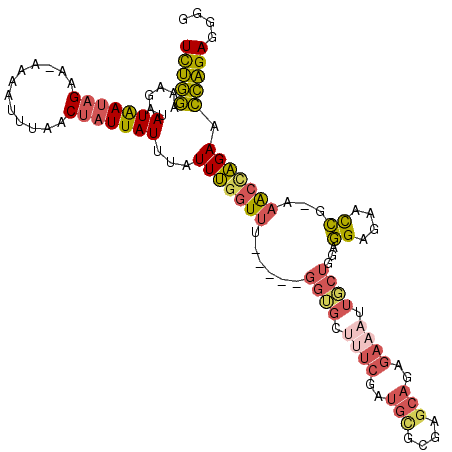
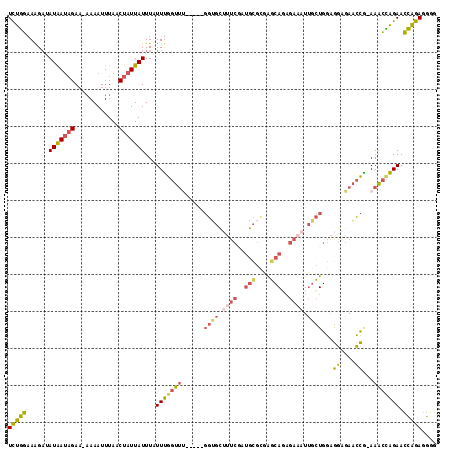
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:49 2006