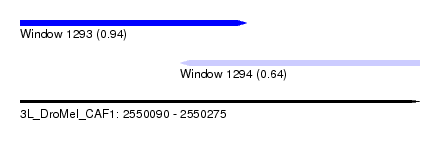
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,550,090 – 2,550,275 |
| Length | 185 |
| Max. P | 0.939349 |

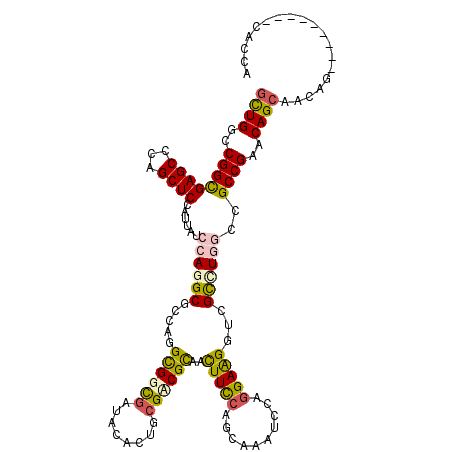
| Location | 2,550,090 – 2,550,195 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.79 |
| Mean single sequence MFE | -59.13 |
| Consensus MFE | -36.22 |
| Energy contribution | -36.33 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2550090 105 + 23771897 GCUGGAGUUGCGCCUGGGCCUGGGCAGCGGCAUGAGCGGCAGCCGCCUGAGCCUGCGCCUGGGCCAUCAGGUGAUGCUG---CUGCUGCUGCUCGGCGGCCAGGCGAC ..........(((((((.((((((((((((((...((((((..(((((((((((......))))..))))))).)))))---)))))))))))))).).))))))).. ( -68.50) >DroVir_CAF1 15656 102 + 1 GAUGCAGCUGC------GCCUGGGCGGCGGCAUGCGCGGCAGCGGCCUGUGCCUGCGCCUGGGCGAGCAGCUGAUGGUGCUGCUGUUGCUGUUCGGCGGCUAGGCGUC (((((((((((------(((..((((.(((...(((((((....))).)))))))))))..)))(((((((.(((((.....)))))))))))).))))))..))))) ( -58.90) >DroGri_CAF1 14289 102 + 1 GAUGUAGCUGC------GCCUGGGCAGCAGCAUGAGCUGCAGUGGCCUGCGCCUGAGCCUGGGCAAGCAACUGAUGAUGCUGCUGCUGCUGUUCGGCGGCGAGGCGAC ......(((.(------(((..(((((((((((.(....((((.((.(((.((.......))))).)).)))).).)))))))))))(((....))))))).)))... ( -50.20) >DroEre_CAF1 25819 105 + 1 GCUGGAGUUGCGCCUGAGCCUGGGCAGCGGCAUGGGCGGCAGCCGCCUGGGCCUGCGCCUGGGCCAUCAGGUGAUGCUG---CUGCUGCUGCUCGGCGGCCAGGCGAC ..........((((((.(((((((((((((((...((((((..(((((((((((......)))))..)))))).)))))---)))))))))))))..))))))))).. ( -65.80) >DroWil_CAF1 22465 99 + 1 GUUGAAGUUGCGC------CUGGGCAGCAGCAUGAACAGCGGCCGCUUGGGCCUGGGCUUGGACCAUCAAUUGAUGGUG---CUGUUGCUGUUCGGCAGCCAAACGAC ((((..(((((((------....)).))))).(((((((((((.(((..(((....)))..)((((((....)))))))---).))))))))))).))))........ ( -45.60) >DroYak_CAF1 10137 105 + 1 GCUGGAGUUGCGCCUGUGCCUGGGCAGCGGCAUGGGCGGCAGCCGCCUGGGCCUGCGCCUGGGCCAUCAGGUGAUGCUG---CUGCUGCUGCUCGGCGGCCAGGCGAC ..........((((((.(((((((((((((((...((((((..(((((((((((......)))))..)))))).)))))---)))))))))))))..))))))))).. ( -65.80) >consensus GCUGGAGUUGCGC____GCCUGGGCAGCGGCAUGAGCGGCAGCCGCCUGGGCCUGCGCCUGGGCCAUCAGGUGAUGCUG___CUGCUGCUGCUCGGCGGCCAGGCGAC (((...(((((.(........).)))))(((.(((((((((((.((((..((....))..))))((((....))))........)))))))))))...))).)))... (-36.22 = -36.33 + 0.12)

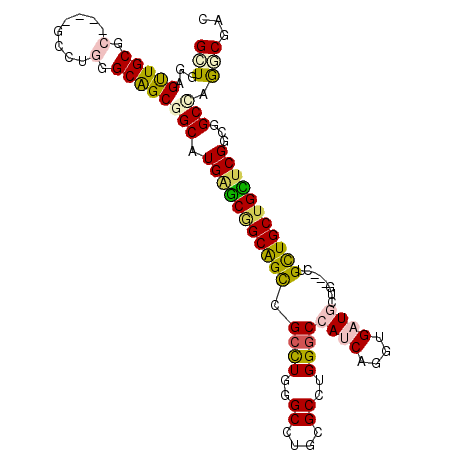
| Location | 2,550,164 – 2,550,275 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -44.58 |
| Consensus MFE | -33.92 |
| Energy contribution | -33.82 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2550164 111 - 23771897 GCUGGCCGGUGAGCACAGCUCCAUUGUCCAGGCACCAGGCGGCGAUACGUUGCGCCGCAACUUCCAGCAGAUCCAGGAAGGUCGCCUGGCCGCCGAGCAGCAGCAG---------CAGCA ((((..((((((((...))))....(.(((((((((..((((((........))))))...((((..........))))))).)))))).)))))..)))).((..---------..)). ( -47.70) >DroVir_CAF1 15724 114 - 1 GCUGGCCGGCGAGCCCAGCUCCAUCAUACAGGCGCCCGGCGGCGAUACAUUGCGUCGCAAUUUCCAGCAAAUCCAGGAGGGACGCCUAGCCGCCGAACAGCAACAGC------AGCACCA ((((..((((((((...))))........((((((((.((((((........))))))....(((..........)))))).)))))....))))..))))......------....... ( -45.90) >DroGri_CAF1 14357 114 - 1 GCUGGCCGGUGAGCCCAGCUCGAUCAUACAAGCGCCUGGUGGCGACACGUUGCGACGCAACUUUCAGCAAAUCCAGGAAGGUCGCCUCGCCGCCGAACAGCAGCAGC------AGCAUCA ((((..(((((.(((((((.(..........).).)))).((((((...((((...))))(((((..........)))))))))))..)))))))..)))).((...------.)).... ( -39.50) >DroWil_CAF1 22533 111 - 1 AUUGGCCGGCGAGCCAAGCUCCAUUAUCCAGGCACCAGGUGGUGAUACCCUGAGACGUAACUUCCAGCAGAUCCAGGAAGGUCGUUUGGCUGCCGAACAGCAACAG---------CACCA ..((..((((.((((((((.........((((((((....))))....)))).(((....(((((..........))))))))))))))))))))..))((....)---------).... ( -39.50) >DroMoj_CAF1 15071 120 - 1 GCUGGCCGGUGAGCCCAGCUCCAUUAUCCAGGCGCCGGGUGGCGAUACACUGCGUCGCAACUUCCAGCAAAUCCAAGAGGGACGCAUGGCCGCCGAACAGCAACAGCAGCAGCAGCACCA ((((..((((..((((.((.((........)).)).))))(((.(.....((((((((........))....((....))))))))).)))))))..))))....((....))....... ( -43.10) >DroAna_CAF1 12463 111 - 1 GCUGGCCGGCGAGCACAGCUCCAUUGUCCAGGCCCCAGGCGGCGACACCCUCCGCCGCAAUUUUCAGCAGAUCCAGGAGGGCCGCCUGGCCGCCGAACAGCAGCAG---------CAGCA ((((..((((((((...)))).........((((..(((((((.....(((((.(.((........)).).....))))))))))))))))))))..)))).((..---------..)). ( -51.80) >consensus GCUGGCCGGCGAGCCCAGCUCCAUUAUCCAGGCGCCAGGCGGCGAUACACUGCGACGCAACUUCCAGCAAAUCCAGGAAGGUCGCCUGGCCGCCGAACAGCAACAG_________CACCA ((((..((((((((...))))......((((((.....((((((........))))))..(((((..........)))))...))))))..))))..))))................... (-33.92 = -33.82 + -0.11)
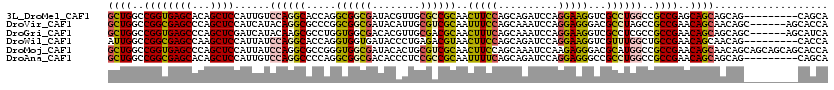
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:30 2006