
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,099,466 – 22,099,669 |
| Length | 203 |
| Max. P | 0.999756 |

| Location | 22,099,466 – 22,099,574 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -25.05 |
| Energy contribution | -26.38 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22099466 108 + 23771897 AACGCCAGCGCCACC------GCCACCGC--CGCAAUAAUCCUGCAGCUCAGUCGAAAUGUCGCUGGCAAUCUAAGGAAAAGGAAAAAUGCCAGCGACAGCGGUGGGGAGUGGGUA .......((.(((((------.(((((((--.(((.......))).............(((((((((((.(((........)))....)))))))))))))))))).).)))))). ( -47.30) >DroSec_CAF1 85479 102 + 1 AACGCCAGCACCACC------GCCACCGC--CGCAAUAAUCCUGCAGCUCAGUCGAAAUGUCGCUGGCAAUCUAAGGAAGAGGAAAAAUGCCAGCGACAGCGGUGGGGAG------ .............((------.(((((((--.(((.......))).............(((((((((((.(((........)))....))))))))))))))))))))..------ ( -40.40) >DroSim_CAF1 91957 102 + 1 AACGCCAGCACCACC------GCCACCGC--CGCAAUAAUCCUGCAGCUCAGUCGAAAUGUCGCUGGCAAUCUAAGGAAGAGGAAAAAUGCCAGCGACAGCGGUGGGGAG------ .............((------.(((((((--.(((.......))).............(((((((((((.(((........)))....))))))))))))))))))))..------ ( -40.40) >DroEre_CAF1 84735 99 + 1 ---GCUACCGCCACC------GCCACCGC--CGCAAUAAUCCUGCAGCUCAGUCGAAAUGUCUCUGGCAAUCUAAGGAAGAGGAAAAAUGCCAACGACAGCGGUGAGGAG------ ---....((..((((------((....((--.(((.......))).))..........((((..(((((.(((........)))....)))))..)))))))))).))..------ ( -29.70) >DroYak_CAF1 103362 93 + 1 ---GC------CACC------GCUAGCAC--CGCAAUAAUCCUGCAGCUGAGUCGAAAUGUCUCUGGCAAUCUAAGGAAGAGGAAAAAUGCCAGCGACAGCGGUGGGGAG------ ---.(------((((------((((((..--.(((.......))).))))........((((.((((((.(((........)))....)))))).)))))))))))....------ ( -34.30) >DroPer_CAF1 104703 113 + 1 CGUGCCUGUGCCACACACUCAGCCA-UGCCGCACAAUAAUCCUGCAACUCAGUCGAAAUGUCGCUGGCAAUCUAAGAAAGAAGAAAAAUGCCAGCAACAUUGGUAGUGAAGGAG-- .((((..(((.....)))...((..-.)).)))).....((((...((((......(((((.(((((((.(((........)))....))))))).))))).).)))..)))).-- ( -28.80) >consensus AACGCCAGCGCCACC______GCCACCGC__CGCAAUAAUCCUGCAGCUCAGUCGAAAUGUCGCUGGCAAUCUAAGGAAGAGGAAAAAUGCCAGCGACAGCGGUGGGGAG______ ......................(((((((...(((.......))).............(((((((((((.(((........)))....)))))))))))))))))).......... (-25.05 = -26.38 + 1.34)

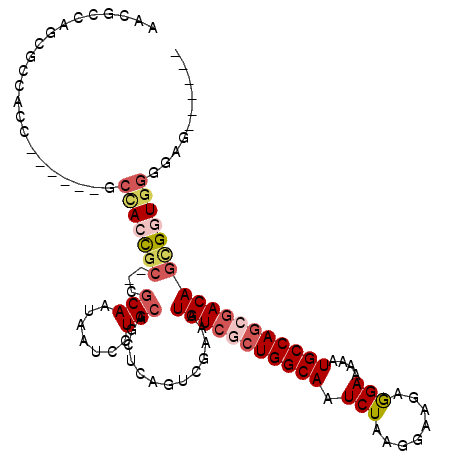

| Location | 22,099,466 – 22,099,574 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -42.35 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22099466 108 - 23771897 UACCCACUCCCCACCGCUGUCGCUGGCAUUUUUCCUUUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGGAUUAUUGCG--GCGGUGGC------GGUGGCGCUGGCGUU .(((((....((((((((((((((((((.(((...........))).))))))))))...........(((((((.....)))))--))...)))------)))))...))).)). ( -48.30) >DroSec_CAF1 85479 102 - 1 ------CUCCCCACCGCUGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGGAUUAUUGCG--GCGGUGGC------GGUGGUGCUGGCGUU ------....((((((((((((((((((.(((...........))).))))))))))...........(((((((.....)))))--))...)))------))))).......... ( -45.70) >DroSim_CAF1 91957 102 - 1 ------CUCCCCACCGCUGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGGAUUAUUGCG--GCGGUGGC------GGUGGUGCUGGCGUU ------....((((((((((((((((((.(((...........))).))))))))))...........(((((((.....)))))--))...)))------))))).......... ( -45.70) >DroEre_CAF1 84735 99 - 1 ------CUCCUCACCGCUGUCGUUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGAGACAUUUCGACUGAGCUGCAGGAUUAUUGCG--GCGGUGGC------GGUGGCGGUAGC--- ------(((((((((((((((.((((((.(((...........))).)))))).)))...........(((((((.....)))))--))...)))------))))).)).)).--- ( -37.30) >DroYak_CAF1 103362 93 - 1 ------CUCCCCACCGCUGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGAGACAUUUCGACUCAGCUGCAGGAUUAUUGCG--GUGCUAGC------GGUG------GC--- ------....(((((((((((.((((((.(((...........))).)))))).)))...........(((((((.....)))))--))...)))------))))------).--- ( -37.50) >DroPer_CAF1 104703 113 - 1 --CUCCUUCACUACCAAUGUUGCUGGCAUUUUUCUUCUUUCUUAGAUUGCCAGCGACAUUUCGACUGAGUUGCAGGAUUAUUGUGCGGCA-UGGCUGAGUGUGUGGCACAGGCACG --.((((.((((...(((((((((((((....(((........))).))))))))))))).......)).)).))))...((((((.(((-(........)))).))))))..... ( -39.60) >consensus ______CUCCCCACCGCUGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGGAUUAUUGCG__GCGGUGGC______GGUGGCGCUGGCGUU ..........(((((..(((((((((((.(((...........))).)))))))))))...(.((((...(((((.....)))))...)))).).......))))).......... (-30.40 = -30.77 + 0.37)



| Location | 22,099,498 – 22,099,603 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.31 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -16.66 |
| Energy contribution | -17.98 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22099498 105 + 23771897 CCUGCAGCUCAGUCGAAAUGUCGCUGGCAAUCUAAGGAAAAGGAAAAAUGCCAGCGACAGCGGUGGGGAGUGGGUAGGCGGUGGCGUGCCCCACCAGUCUGUUCA ((..(..((((.(((...(((((((((((.(((........)))....))))))))))).)))))))..)..))((((((((((......))))).))))).... ( -45.70) >DroPse_CAF1 103583 82 + 1 CCUGCAACUCAGUCGAAAUGUCGCUGGCAAUCUAAGAAAGAAGAAAAAUGCCAGCAACAUUGGUAGUGAAGGAG-----------------------GAGGAGCA .((.(..(((..(((.(((((.(((((((.(((........)))....))))))).))))).....)))..)))-----------------------..).)).. ( -25.70) >DroEre_CAF1 84764 95 + 1 CCUGCAGCUCAGUCGAAAUGUCUCUGGCAAUCUAAGGAAGAGGAAAAAUGCCAACGACAGCGGUGAGGAG----------CUGGCGUGCCCCACCAGUCAGUUCA .......((((.(((...((((..(((((.(((........)))....)))))..)))).)))))))(((----------((((((((...)))..)))))))). ( -30.00) >DroYak_CAF1 103385 95 + 1 CCUGCAGCUGAGUCGAAAUGUCUCUGGCAAUCUAAGGAAGAGGAAAAAUGCCAGCGACAGCGGUGGGGAG----------AUGGCGUGCCCCACCAGUCAGUUCA ...(.((((((...(...((((.((((((.(((........)))....)))))).)))).)(((((((.(----------....)...)))))))..))))))). ( -35.80) >DroPer_CAF1 104742 82 + 1 CCUGCAACUCAGUCGAAAUGUCGCUGGCAAUCUAAGAAAGAAGAAAAAUGCCAGCAACAUUGGUAGUGAAGGAG-----------------------GAGGAGCA .((.(..(((..(((.(((((.(((((((.(((........)))....))))))).))))).....)))..)))-----------------------..).)).. ( -25.70) >consensus CCUGCAGCUCAGUCGAAAUGUCGCUGGCAAUCUAAGGAAGAGGAAAAAUGCCAGCGACAGCGGUGGGGAG_G_G_______UGGCGUGCCCCACCAGUCAGUUCA .......(((..(((...(((((((((((.(((........)))....))))))))))).)))..)))..................................... (-16.66 = -17.98 + 1.32)

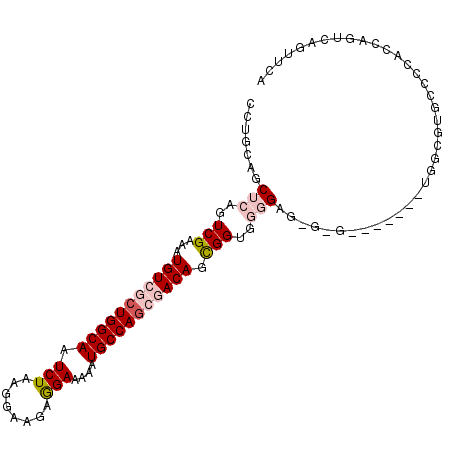
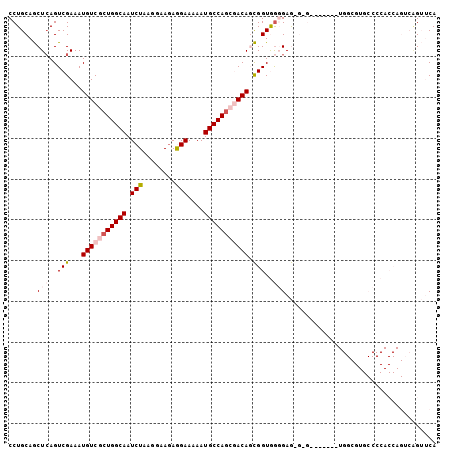
| Location | 22,099,498 – 22,099,603 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.31 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -19.09 |
| Energy contribution | -22.01 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22099498 105 - 23771897 UGAACAGACUGGUGGGGCACGCCACCGCCUACCCACUCCCCACCGCUGUCGCUGGCAUUUUUCCUUUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGG ....(((...(((((((....................)))))))..(((((((((((.(((...........))).)))))))))))......)))......... ( -35.85) >DroPse_CAF1 103583 82 - 1 UGCUCCUC-----------------------CUCCUUCACUACCAAUGUUGCUGGCAUUUUUCUUCUUUCUUAGAUUGCCAGCGACAUUUCGACUGAGUUGCAGG (((..(((-----------------------.............(((((((((((((....(((........))).)))))))))))))......)))..))).. ( -25.11) >DroEre_CAF1 84764 95 - 1 UGAACUGACUGGUGGGGCACGCCAG----------CUCCUCACCGCUGUCGUUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGAGACAUUUCGACUGAGCUGCAGG ....(((...((((.....))))((----------(((........((((.((((((.(((...........))).)))))).))))........))))).))). ( -27.19) >DroYak_CAF1 103385 95 - 1 UGAACUGACUGGUGGGGCACGCCAU----------CUCCCCACCGCUGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGAGACAUUUCGACUCAGCUGCAGG ((..((((..(((((((........----------..)))))))..((((.((((((.(((...........))).)))))).)))).......))))...)).. ( -31.40) >DroPer_CAF1 104742 82 - 1 UGCUCCUC-----------------------CUCCUUCACUACCAAUGUUGCUGGCAUUUUUCUUCUUUCUUAGAUUGCCAGCGACAUUUCGACUGAGUUGCAGG (((..(((-----------------------.............(((((((((((((....(((........))).)))))))))))))......)))..))).. ( -25.11) >consensus UGAACCGACUGGUGGGGCACGCCA_______C_C_CUCCCCACCGCUGUCGCUGGCAUUUUUCCUCUUCCUUAGAUUGCCAGCGACAUUUCGACUGAGCUGCAGG ..........(((((((....................)))))))..(((((((((((.(((...........))).)))))))))))......(((.....))). (-19.09 = -22.01 + 2.92)

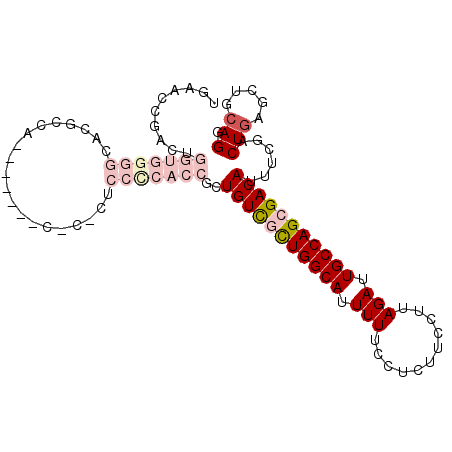
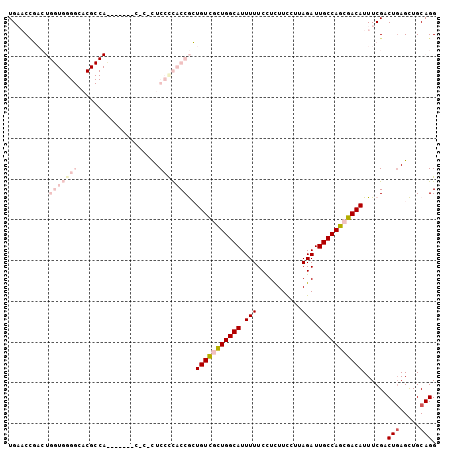
| Location | 22,099,536 – 22,099,637 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 70.09 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -18.80 |
| Energy contribution | -21.72 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.54 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22099536 101 + 23771897 AAAGGAAAAAUGCCAGCGACAGCGGUGGGGAGUGGGUAGGCGGUGGCGUGCCCCACCAGUCUGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAAC ...((.......))((((((((((((..((.(.(((((.((....)).)))))).)).(((((((..(((.....)))))))))))))))))))))).... ( -45.90) >DroPse_CAF1 103621 78 + 1 AGAAGAAAAAUGCCAGCAACAUUGGUAGUGAAGGAG-----------------------GAGGAGCAUCUUCUGGAGCAGCAUCUGCUGCUGUCGCUCCAA .....................((((.(((((.((((-----------------------((......))))))..((((((....)))))).))))))))) ( -24.70) >DroEre_CAF1 84802 91 + 1 AGAGGAAAAAUGCCAACGACAGCGGUGAGGAG----------CUGGCGUGCCCCACCAGUCAGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAAC .((((........)..(((((((((((.....----------(((.(.(((.((((((((.......)))))))).))))))).))))))))))))))... ( -37.70) >DroYak_CAF1 103423 91 + 1 AGAGGAAAAAUGCCAGCGACAGCGGUGGGGAG----------AUGGCGUGCCCCACCAGUCAGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAAC ...((.......))(((((((((((((.....----------...((.(((.((((((((.......)))))))).)))))...))))))))))))).... ( -41.50) >DroPer_CAF1 104780 78 + 1 AGAAGAAAAAUGCCAGCAACAUUGGUAGUGAAGGAG-----------------------GAGGAGCAUCUUCUGGAGCAGCAUCUGCUGCUGUCGCUCCAA .....................((((.(((((.((((-----------------------((......))))))..((((((....)))))).))))))))) ( -24.70) >consensus AGAGGAAAAAUGCCAGCGACAGCGGUGGGGAG_G_G_______UGGCGUGCCCCACCAGUCAGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAAC ..............((((((................................((((((((.......))))))))((((((....)))))))))))).... (-18.80 = -21.72 + 2.92)

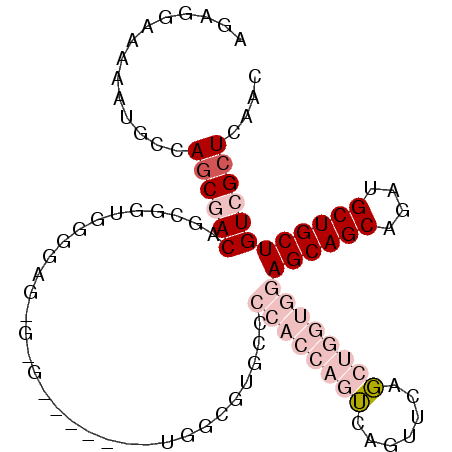
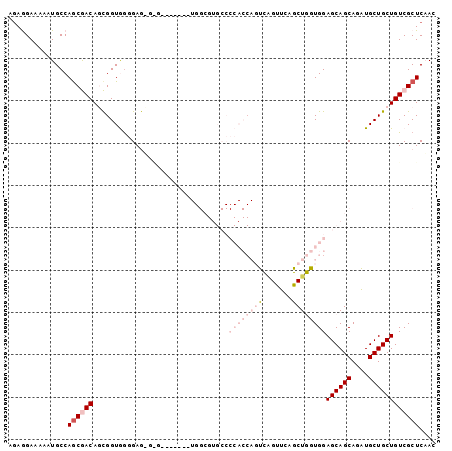
| Location | 22,099,536 – 22,099,637 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 70.09 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -20.24 |
| Energy contribution | -22.16 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.62 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22099536 101 - 23771897 GUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACAGACUGGUGGGGCACGCCACCGCCUACCCACUCCCCACCGCUGUCGCUGGCAUUUUUCCUUU (((.((((((((((((....)))((((((((((((....)).))))))))))..........................))))))))))))........... ( -42.70) >DroPse_CAF1 103621 78 - 1 UUGGAGCGACAGCAGCAGAUGCUGCUCCAGAAGAUGCUCCUC-----------------------CUCCUUCACUACCAAUGUUGCUGGCAUUUUUCUUCU ....((((((((((((....)))))....((((..(......-----------------------)..))))........))))))).............. ( -20.00) >DroEre_CAF1 84802 91 - 1 GUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACUGACUGGUGGGGCACGCCAG----------CUCCUCACCGCUGUCGUUGGCAUUUUUCCUCU ((..(.((((((((((....)))(((((((((((......).)))))))))).......----------.........))))))))..))........... ( -39.70) >DroYak_CAF1 103423 91 - 1 GUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACUGACUGGUGGGGCACGCCAU----------CUCCCCACCGCUGUCGCUGGCAUUUUUCCUCU (((.((((((((((((....)))(((((((((((......).)))))))))).......----------.........))))))))))))........... ( -41.60) >DroPer_CAF1 104780 78 - 1 UUGGAGCGACAGCAGCAGAUGCUGCUCCAGAAGAUGCUCCUC-----------------------CUCCUUCACUACCAAUGUUGCUGGCAUUUUUCUUCU ....((((((((((((....)))))....((((..(......-----------------------)..))))........))))))).............. ( -20.00) >consensus GUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACCGACUGGUGGGGCACGCCA_______C_C_CUCCCCACCGCUGUCGCUGGCAUUUUUCCUCU ....((((((((((((....))))).((((((((......).)))))))...............................))))))).............. (-20.24 = -22.16 + 1.92)

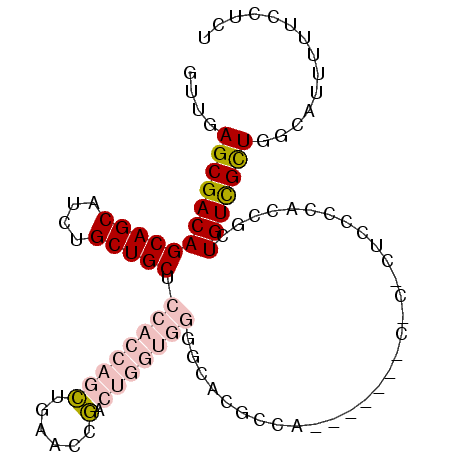

| Location | 22,099,574 – 22,099,669 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22099574 95 + 23771897 GGCGGUGGCGUGCCCCACCAGUCUGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAACCACCCACUUCCA--CCUCCUCAACUCCUCCGCCG (((((.((.((...(((((((.((....)))))))))((((((....))))))......................--........)).)).))))). ( -39.50) >DroSec_CAF1 85581 76 + 1 ---------------------UCAGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAACCACCCACUUCCACACCUCCUCCACUCCUCCGCCG ---------------------...........(((((((((((....)))))((.....)).............................)))))). ( -17.40) >DroSim_CAF1 92059 76 + 1 ---------------------UCAGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAACCACCCACUUCCACACCUCCUCCACUCCGCCGCCG ---------------------........((.(((((((((((....)))))((.....))..........................)))))))).. ( -18.70) >DroEre_CAF1 84834 92 + 1 ----CUGGCGUGCCCCACCAGUCAGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAACCACCCACUUCCACACCUCGUACACCACAUCC-CC ----.(((.((((.((((((((.......))))))))((((((....))))))............................)))).))).....-.. ( -31.30) >DroYak_CAF1 103455 85 + 1 ----AUGGCGUGCCCCACCAGUCAGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAACCACCCACUUCCACACCUCGUCCACCG-------- ----.(((.(((..((((((((.......))))))))((((((....))))))..........))))))....................-------- ( -28.80) >consensus _____UGGCGUGCCCCACCAGUCAGUUCAGCUGGUGGAGCAGCAGAUGCUGCUGUCGCUCAACCACCCACUUCCACACCUCCUCCACUCCUCCGCCG ................................(((((((((((....))))))(.....)..))))).............................. (-16.20 = -16.20 + -0.00)


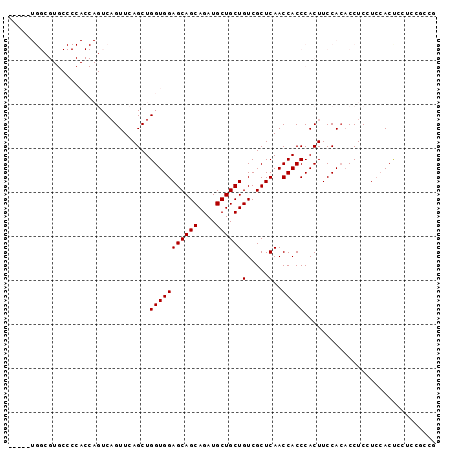
| Location | 22,099,574 – 22,099,669 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22099574 95 - 23771897 CGGCGGAGGAGUUGAGGAGG--UGGAAGUGGGUGGUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACAGACUGGUGGGGCACGCCACCGCC ((((......)))).((.((--(((.((..((((((((.....))))..))))..))((((((((((((....)).))))))))))...))))).)) ( -44.50) >DroSec_CAF1 85581 76 - 1 CGGCGGAGGAGUGGAGGAGGUGUGGAAGUGGGUGGUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACUGA--------------------- ((((((.((((..(...((((((....((..((.(.....).)).)).)))))).)..)))).)).))))......--------------------- ( -27.20) >DroSim_CAF1 92059 76 - 1 CGGCGGCGGAGUGGAGGAGGUGUGGAAGUGGGUGGUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACUGA--------------------- ((((((.((((..(...((((((....((..((.(.....).)).)).)))))).)..)))).)).))))......--------------------- ( -28.00) >DroEre_CAF1 84834 92 - 1 GG-GGAUGUGGUGUACGAGGUGUGGAAGUGGGUGGUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACUGACUGGUGGGGCACGCCAG---- ..-.....((((((............((..((((((((.....))))..))))..))(((((((((((......).)))))))))))))))).---- ( -37.80) >DroYak_CAF1 103455 85 - 1 --------CGGUGGACGAGGUGUGGAAGUGGGUGGUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACUGACUGGUGGGGCACGCCAU---- --------((.....)).(((((...((..((((((((.....))))..))))..))(((((((((((......).)))))))))))))))..---- ( -37.40) >consensus CGGCGGAGGAGUGGAGGAGGUGUGGAAGUGGGUGGUUGAGCGACAGCAGCAUCUGCUGCUCCACCAGCUGAACUGACUGGUGGGGCACGCCA_____ .....................((...(((.(((((.........((((((....))))))))))).)))..))....(((((.....)))))..... (-20.18 = -20.88 + 0.70)

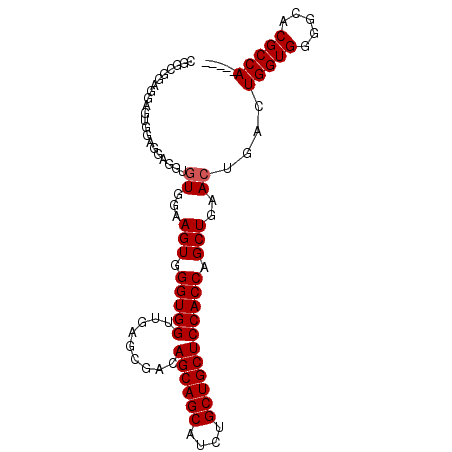

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:38 2006