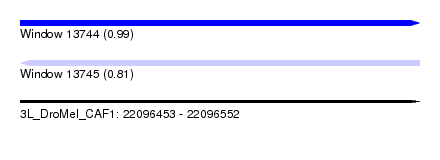
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,096,453 – 22,096,552 |
| Length | 99 |
| Max. P | 0.988467 |

| Location | 22,096,453 – 22,096,552 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.18 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -10.17 |
| Energy contribution | -10.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
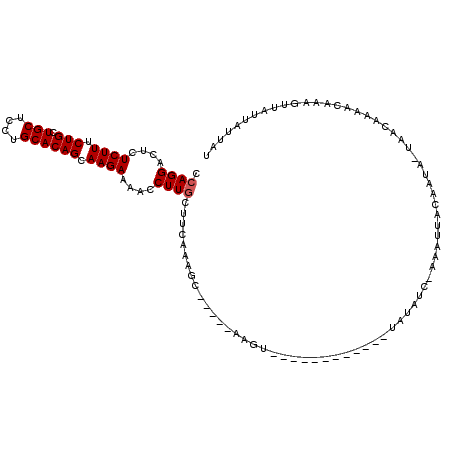
>3L_DroMel_CAF1 22096453 99 + 23771897 CCAGGACUCUCUUUCUGCUGCUUCUGCACAGCAAGAAAACCUUGGUUCAAAGC-----AAGU------------UAUAUCAAAAUUGCAAUGCUAAUAAAACAAUGUUAUUAAUAU (((((....((((.(((.(((....)))))).))))....)))))......((-----((..------------..........)))).....((((((.......)))))).... ( -17.50) >DroSim_CAF1 88889 94 + 1 CCAGGACUCUCUUUCUGCUGCUCCUGCACAGCAAGAAAACCUUGCUUCAAAGC-----AAGU------------UAUAU----AUUAUUAUA-UAACACAGCAAGAUCACUUUUAA .............(((((((....(((..((((((.....)))))).....))-----).((------------(((((----(....))))-)))).)))).))).......... ( -21.60) >DroYak_CAF1 100349 116 + 1 CCAGGACUCUCUUUCUGCUGCUGCUGCACAGUAAGAAAAACUUACUGAAACUCAAAACAAAGUUUAAUUUGGCUUAUAUCGAAAGUACAAUAUUAACAAAACCAAGUUAUUAUUAU ..........(((((.((....)).((.(((((((.....)))))))..........((((......)))))).......)))))...((((.((((........)))).)))).. ( -18.70) >consensus CCAGGACUCUCUUUCUGCUGCUCCUGCACAGCAAGAAAACCUUGCUUCAAAGC_____AAGU____________UAUAUC_AAAUUACAAUA_UAACAAAACAAAGUUAUUAUUAU .((((....((((.(((.(((....)))))).))))....))))........................................................................ (-10.17 = -10.50 + 0.33)


| Location | 22,096,453 – 22,096,552 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.18 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -13.15 |
| Energy contribution | -12.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
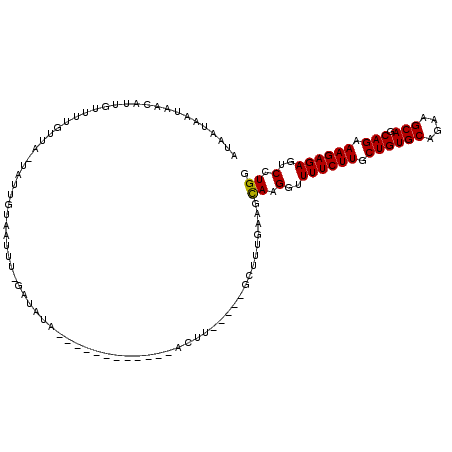
>3L_DroMel_CAF1 22096453 99 - 23771897 AUAUUAAUAACAUUGUUUUAUUAGCAUUGCAAUUUUGAUAUA------------ACUU-----GCUUUGAACCAAGGUUUUCUUGCUGUGCAGAAGCAGCAGAAAGAGAGUCCUGG ...(((((((.......)))))))((..((((.((......)------------).))-----))..))..(((.(..((((((.((((((....))).))).))))))..).))) ( -22.10) >DroSim_CAF1 88889 94 - 1 UUAAAAGUGAUCUUGCUGUGUUA-UAUAAUAAU----AUAUA------------ACUU-----GCUUUGAAGCAAGGUUUUCUUGCUGUGCAGGAGCAGCAGAAAGAGAGUCCUGG .....((.((((((.((((((((-((((....)----)))))------------)).(-----((((((.((((((.....))))))...)).)))))))))...)))).)))).. ( -27.40) >DroYak_CAF1 100349 116 - 1 AUAAUAAUAACUUGGUUUUGUUAAUAUUGUACUUUCGAUAUAAGCCAAAUUAAACUUUGUUUUGAGUUUCAGUAAGUUUUUCUUACUGUGCAGCAGCAGCAGAAAGAGAGUCCUGG .......(((.(((((((.....((((((......))))))))))))).))).(((((.((((..((..(((((((.....))))))).)).((....)).)))).)))))..... ( -24.40) >consensus AUAAUAAUAACAUUGUUUUGUUA_UAUUGUAAUUU_GAUAUA____________ACUU_____GCUUUGAAGCAAGGUUUUCUUGCUGUGCAGAAGCAGCAGAAAGAGAGUCCUGG ........................................................................((.(..((((((.((((((....))).))).))))))..).)). (-13.15 = -12.93 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:31 2006