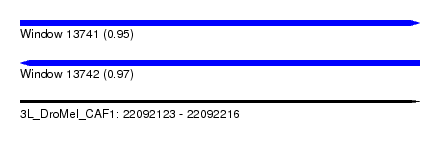
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,092,123 – 22,092,216 |
| Length | 93 |
| Max. P | 0.974552 |

| Location | 22,092,123 – 22,092,216 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -16.41 |
| Energy contribution | -18.54 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
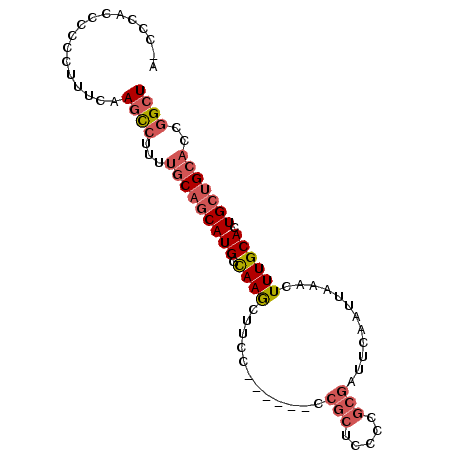
>3L_DroMel_CAF1 22092123 93 + 23771897 AGCCGGUGCAGCAGUGCAAAGUUUAAUUGAAUCGCGGGGAGCGGGGAAGGGGAAGCUUACCAUGCUGCAAAAGGAUGGUCAGGGGGGAGGAAU ..((..(((((((.((..((((((..((...((((.....))))....))..))))))..)))))))))...))................... ( -24.90) >DroSec_CAF1 77690 79 + 1 AGCCGGUGCAGCAGUGCAAAGUUUAAUUGAAUCGCGG-------------GGAAGCUUGACAUGCUGCAAAAGGCUUGAAAGGUGGGUGGG-U ((((..(((((((.((..((((((..(((.....)))-------------..))))))..)))))))))...))))...............-. ( -25.50) >DroSim_CAF1 82611 86 + 1 AGCCGGUGCAGCAGUGCAAAGUUUAAUUGAAUCGCGGGGAGCGG------GGAAGCUUGACAUGCUGCAAAAGGCUUGAAAGGGGGGUGGG-G ((((..(((((((.((..((((((.......((((.....))))------..))))))..)))))))))...))))...............-. ( -28.20) >DroYak_CAF1 92423 76 + 1 AGCCGGUGCAGCAGUGCAAAGUUUAAUUGAAUCGCGGGGAGCGG------GGAAGUUUGCCAUGUGG------ACUGGAAAGG--GGU--G-G ..(((((.((.((..(((((.(((.......((((.....))))------.))).)))))..)))).------))))).....--...--.-. ( -21.40) >consensus AGCCGGUGCAGCAGUGCAAAGUUUAAUUGAAUCGCGGGGAGCGG______GGAAGCUUGACAUGCUGCAAAAGGCUGGAAAGGGGGGUGGG_G ((((..(((((((.((..((((((.......((((.....))))........))))))..)))))))))...))))................. (-16.41 = -18.54 + 2.13)



| Location | 22,092,123 – 22,092,216 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -16.25 |
| Consensus MFE | -10.74 |
| Energy contribution | -12.43 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22092123 93 - 23771897 AUUCCUCCCCCCUGACCAUCCUUUUGCAGCAUGGUAAGCUUCCCCUUCCCCGCUCCCCGCGAUUCAAUUAAACUUUGCACUGCUGCACCGGCU ...................((...(((((((.((..(((............)))..))((((............))))..)))))))..)).. ( -18.90) >DroSec_CAF1 77690 79 - 1 A-CCCACCCACCUUUCAAGCCUUUUGCAGCAUGUCAAGCUUCC-------------CCGCGAUUCAAUUAAACUUUGCACUGCUGCACCGGCU .-...............((((...((((((((((.(((.....-------------................))).))).)))))))..)))) ( -17.80) >DroSim_CAF1 82611 86 - 1 C-CCCACCCCCCUUUCAAGCCUUUUGCAGCAUGUCAAGCUUCC------CCGCUCCCCGCGAUUCAAUUAAACUUUGCACUGCUGCACCGGCU .-...............((((...(((((((.((..(((....------..)))....((((............)))))))))))))..)))) ( -20.50) >DroYak_CAF1 92423 76 - 1 C-C--ACC--CCUUUCCAGU------CCACAUGGCAAACUUCC------CCGCUCCCCGCGAUUCAAUUAAACUUUGCACUGCUGCACCGGCU .-.--...--.......(((------(.....((((.......------.(((.....)))...(((.......)))...)))).....)))) ( -7.80) >consensus A_CCCACCCCCCUUUCAAGCCUUUUGCAGCAUGGCAAGCUUCC______CCGCUCCCCGCGAUUCAAUUAAACUUUGCACUGCUGCACCGGCU .................((((...(((((((((.((((............(((.....)))............)))))).)))))))..)))) (-10.74 = -12.43 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:28 2006