| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,090,314 – 22,090,414 |
| Length | 100 |
| Max. P | 0.566751 |

| Location | 22,090,314 – 22,090,414 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
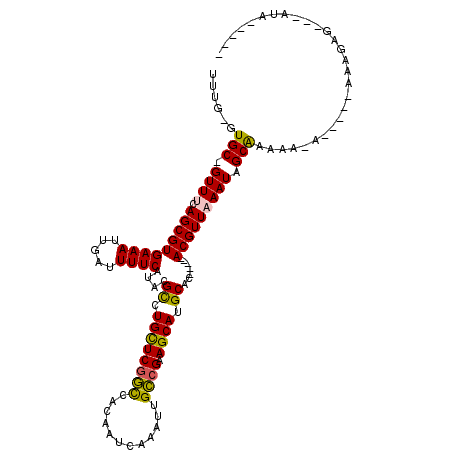
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -21.26 |
| Energy contribution | -20.93 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22090314 100 - 23771897 UUUG-GUGC-GUUUCAGCGUGAAAUUGAUUUUCAUACGCCUGCUCGGCCACAAUCAAAUUGCCGAAGCAUGCAC-----ACGUUUAAUAGCAAAAAAA-----AAAGAG---AUA----- (((.-.(((-.....((((((....((.....))...((.((((((((............)))).)))).)).)-----))))).....)))..))).-----......---...----- ( -24.40) >DroPse_CAF1 92017 109 - 1 UUUG-GUGC-GUUUCAGCGUGAAAUUGAUUUUCAUACGCCUGCUCGGCCACAAUCAAAUUGCCGAAGCAUGCAC-----ACGUUAAAUAGCACAAA-AAA---AAAGAGGAGAUACAUAA ....-((((-((((.((((((....((.....))...((.((((((((............)))).)))).)).)-----))))))))).))))...-...---................. ( -27.10) >DroGri_CAF1 85755 100 - 1 UUUG-GUGC-GUUUCAGCGUGAAAUUGAUUUUCAUACGCCUGCUCGGUCACAAUCAAAUUGCAGAAGCAUGC-------ACGUUAAAUAGCAAAAA-AGCUUCAACGA-----UA----- ....-((((-(((((.((((((((.....))))))..(((.....)))............)).)))))..))-------))(((....(((.....-.)))..)))..-----..----- ( -20.60) >DroSec_CAF1 75940 104 - 1 CUUG-GUGC-GUUUCAGCGUGAAAUUGAUUUUCAUACGUCUGUUCGGCCACAAUCAAAUUGCCGAAGCAUGCACACGACACGUUUAAUAGCAAAAA-A-----AAAGAG---AUA----- .(((-((((-((..((((((((((.....))))..))).)))((((((............))))))..)))))).)))...(((....))).....-.-----......---...----- ( -20.90) >DroAna_CAF1 97404 96 - 1 UUUG-GUGC-GUUUCAGCGUGAAAUUGAUUUUCAUACGCCUGCUCGGCCACAAUCAAAUUGCCGAAGCAUGCAC-----ACGUUAAAUAGCGAAAA-A-----A---GG---AUA----- (((.-.(((-((((.((((((....((.....))...((.((((((((............)))).)))).)).)-----))))))))).)))..))-)-----.---..---...----- ( -24.10) >DroPer_CAF1 92148 110 - 1 UUUGGGUGCGGUUUCAGCGUGAAAUAAAUUUUCAUCCGCAUGCUCGCCCACACUCAAAUUGGCGAAGCAAGCAC-----ACGUUAAAUAGCACAAA-A-A---AAAGAGGAGAUACAUAA (((..((((.((((.(((((((((.....))))....((.((((((((............)))).)))).))..-----))))))))).))))..)-)-)---................. ( -28.10) >consensus UUUG_GUGC_GUUUCAGCGUGAAAUUGAUUUUCAUACGCCUGCUCGGCCACAAUCAAAUUGCCGAAGCAUGCAC_____ACGUUAAAUAGCAAAAA_A_____AAAGAG___AUA_____ ......(((.((((.(((((((((.....))))....((.((((((((............)))).)))).)).......))))))))).)))............................ (-21.26 = -20.93 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:27 2006