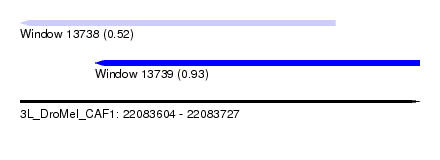
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,083,604 – 22,083,727 |
| Length | 123 |
| Max. P | 0.927049 |

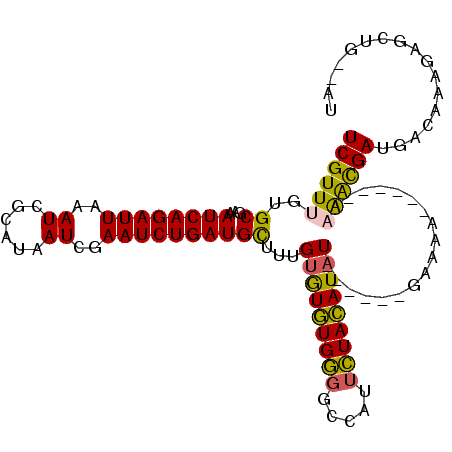
| Location | 22,083,604 – 22,083,701 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -17.53 |
| Energy contribution | -16.37 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22083604 97 - 23771897 UCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGUUUAGUGUGUGGGGCCAUUCUACAUAU------GAAA-------AAAACGAUGACAAAGAGCUG--AU (((.((((((((............)))))))).))).(((..((((..(((((((((.....)))))))))------....-------........)))).)))....--.. ( -21.14) >DroPse_CAF1 84417 108 - 1 UCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUACGUAUAUGU-ACGACUAUAUAUAUGUAUAUGUAUAG---AGUCGAUGAUGACAAAGACCUGGGAU ((((((((((((............)))))))(((((.((((.((((((((((((((-(....))))))))))))...))))))---).)))))))))).............. ( -27.90) >DroSec_CAF1 69127 100 - 1 UCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUAU------GAAAAA----AAAAACGAUGACAAAGAGCUC--AU (((.((((((((............)))))))).)))...(((.((((((((((((((.....)))))))))------......----......(....)..)))))))--). ( -22.60) >DroEre_CAF1 69254 104 - 1 UCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUUU------GAAAAAACGAAAAAACGAUGACAAAGAGCUG--AU (((.((((((((............)))))))).)))(((.(...(((((((((((((.....)))))))..------.......((......))...))))))..).)--)) ( -22.50) >DroYak_CAF1 84022 98 - 1 UCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUAU------GAAAA------AAAGCGAUGACAAAGAGCUG--AU ((((....)))).(((((.....(((((...((((....))))..((..((((((((.....)))))))).------.))..------...))))).........)))--)) ( -23.34) >DroPer_CAF1 84107 103 - 1 UCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUACGUAUAUGUGACGACUAUAUAUAU------GUAUAG---AGUCGAUGAUGACAAAGACCUGGGAU (((.((((((((............)))))))).)))((((.....((((((((((((.....)))))))))------)))(((---.(((..((....))..)))))))))) ( -26.70) >consensus UCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUAU______GAAAA______AAAACGAUGACAAAGAGCUG__AU ((((((..((...((((((((..((......))..))))))))))...(((((((((.....)))))))))..................))))))................. (-17.53 = -16.37 + -1.16)

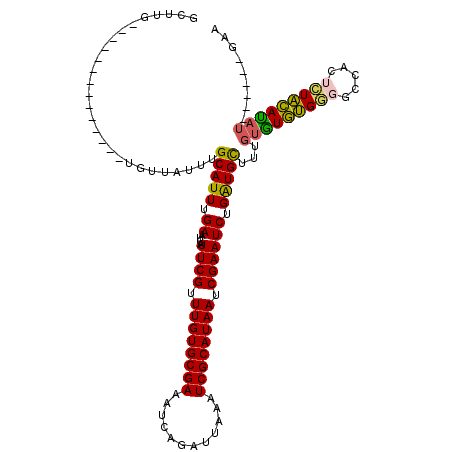
| Location | 22,083,627 – 22,083,727 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -22.98 |
| Energy contribution | -22.17 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22083627 100 - 23771897 GCUUG--------------UAUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGUUUAGUGUGUGGGGCCAUUCUACAUAU------GAA ...((--------------((.....)))).....((((((((.((((((((............)))))))).))))..)))).....(((((((((.....)))))))))------... ( -23.20) >DroPse_CAF1 84446 105 - 1 GUUUG--------------UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUACGUAUAUGU-ACGACUAUAUAUAUGUAUAUGUA .....--------------........(((((.((....((((.((((((((............)))))))).)))))).)))))(((((((((((-(....))))))))))))...... ( -27.30) >DroEre_CAF1 69284 100 - 1 GCUUG--------------UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUUU------GAA .....--------------........(((((.((....((((.((((((((............)))))))).)))))).))))).....(((((((.....)))))))..------... ( -22.20) >DroYak_CAF1 84046 100 - 1 GCUUG--------------UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCAUUCUACAUAU------GAA .....--------------........(((((.((....((((.((((((((............)))))))).)))))).))))).(..((((((((.....)))))))).------.). ( -26.40) >DroAna_CAF1 90797 105 - 1 GCCGGGUGGUGCGGUGGCCUGGUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAGUCGCAUAAUCGAAUCUGGUGCUUUAUGUGC-----CGCUC-GCACAC--------- ........((((((((((...(((...((((..((....((((.((((((((............)))))))).))))))..))))..)))..))-----))).)-))))..--------- ( -36.40) >DroPer_CAF1 84136 100 - 1 GUUUG--------------UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUACGUAUAUGUGACGACUAUAUAUAU------GUA .....--------------........(((((.((....((((.((((((((............)))))))).)))))).)))))((((((((((((.....)))))))))------))) ( -26.70) >consensus GCUUG______________UGUUAUUUGCAUUUGAAUCAUUCGUUUGUGCGAAAUCAGAUUAAAUCGCAUAAUCGAAUCUGAUGCUUUGUGUGUGGGGCCACUCUACAUAU______GAA ...........................(((((.((....((((.((((((((............)))))))).)))))).)))))...(((((((((.....)))))))))......... (-22.98 = -22.17 + -0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:26 2006