| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,082,306 – 22,082,400 |
| Length | 94 |
| Max. P | 0.850852 |

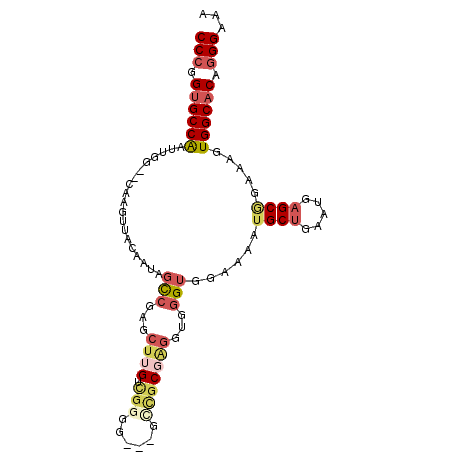
| Location | 22,082,306 – 22,082,400 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -24.19 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22082306 94 + 23771897 UUUCCCUGUGCCACUUUCCGCUCAUUCAGCAUUUCCCACCCACCCCGCGGC---CCCCGGCCAGCUCGGCUUUUGUAACUUG--CCAAUUGGCACCGGG ...(((.((((((......(((.....)))................(((((---(...)))).))..(((...........)--))...)))))).))) ( -28.50) >DroSec_CAF1 67850 95 + 1 UUUCCCUGUGCCACUUUCUGCUCAUUCAGCAUUUUCCACCCACCCCGCGAC---CCCCGACAAGC-CGGCUAUUGUAACUUGCGCCAAUUGGCACCGGG ...(((.((((((.....((((.....))))...............(((..---.....((((..-......))))......)))....)))))).))) ( -23.12) >DroSim_CAF1 74206 96 + 1 UUUCCCUGUGCCACUUUCUGCUCAUUCAGCAUUUUCCACCCACCCCGCGAC---CCCCGACAAGCCCGGCUAUUGUAACUUGCGCCAAUUGGCACCGGG ...(((.((((((.....((((.....))))...............(((..---.....((((((...)))..)))......)))....)))))).))) ( -22.72) >DroEre_CAF1 67983 96 + 1 UUUCCCUGUGCCACUUUCCGCUCAUUCAGCUUUUUCCACCCACCUCGCCCCCUCCCCCGACAAGC-UGGCUAUUGUAACUUG--CCAAUUGGCACCGGG ...(((.((((((......(((.....)))...................................-((((...........)--)))..)))))).))) ( -21.10) >DroYak_CAF1 82751 92 + 1 UUUCCCUGUGCCACUUUCCGCUCAUUCUGCAUUUUUCACCCACCUCGCAGC-----CCACCAAGCUUGGCUAUUGUAACUUG--CCAAUUGGCACCGGG ...(((.((((((......(((....((((................)))).-----......)))(((((...........)--)))).)))))).))) ( -24.59) >DroAna_CAF1 89422 80 + 1 CCUCCGCGGGCCACUUGCCGCUUAUUCAGCAUUUCGGUCGGGCAUCGCAGC---UCUC--C--GUUCGAC----------UG--CCAAUCGGCACCGGG ...((.((((((.......(((.....))).....(((..(((((((.(((---....--.--)))))).----------))--)).)))))).))))) ( -25.10) >consensus UUUCCCUGUGCCACUUUCCGCUCAUUCAGCAUUUUCCACCCACCCCGCGGC___CCCCGACAAGCUCGGCUAUUGUAACUUG__CCAAUUGGCACCGGG ...(((.(((((.......(((.....)))..........................(((.......))).....................))))).))) (-14.59 = -14.70 + 0.11)

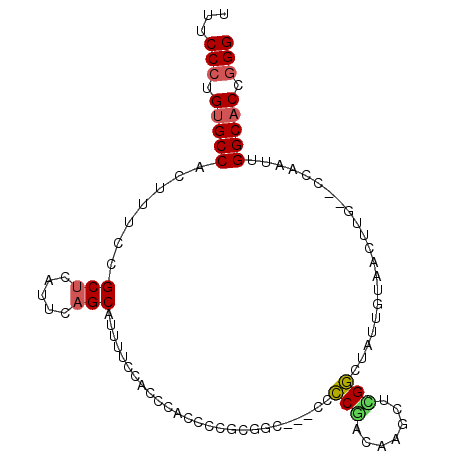
| Location | 22,082,306 – 22,082,400 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -18.60 |
| Energy contribution | -19.93 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22082306 94 - 23771897 CCCGGUGCCAAUUGG--CAAGUUACAAAAGCCGAGCUGGCCGGGG---GCCGCGGGGUGGGUGGGAAAUGCUGAAUGAGCGGAAAGUGGCACAGGGAAA (((.(((((..(..(--((..(..((...(((..((.((((...)---)))))..)))...))..)..)))..)....((.....))))))).)))... ( -37.00) >DroSec_CAF1 67850 95 - 1 CCCGGUGCCAAUUGGCGCAAGUUACAAUAGCCG-GCUUGUCGGGG---GUCGCGGGGUGGGUGGAAAAUGCUGAAUGAGCAGAAAGUGGCACAGGGAAA (((.((((((..((((....))))...((.(((-.((((.((...---..)))))).))).)).....((((.....)))).....)))))).)))... ( -32.90) >DroSim_CAF1 74206 96 - 1 CCCGGUGCCAAUUGGCGCAAGUUACAAUAGCCGGGCUUGUCGGGG---GUCGCGGGGUGGGUGGAAAAUGCUGAAUGAGCAGAAAGUGGCACAGGGAAA (((.((((((..((((....)))).....(((.(.((((.((...---..)))))).).)))......((((.....)))).....)))))).)))... ( -32.90) >DroEre_CAF1 67983 96 - 1 CCCGGUGCCAAUUGG--CAAGUUACAAUAGCCA-GCUUGUCGGGGGAGGGGGCGAGGUGGGUGGAAAAAGCUGAAUGAGCGGAAAGUGGCACAGGGAAA (((.((((((.((.(--(.((((....((.(((-.((((((.........)))))).))).)).....))))......)).))...)))))).)))... ( -33.20) >DroYak_CAF1 82751 92 - 1 CCCGGUGCCAAUUGG--CAAGUUACAAUAGCCAAGCUUGGUGG-----GCUGCGAGGUGGGUGAAAAAUGCAGAAUGAGCGGAAAGUGGCACAGGGAAA (((.((((((.((((--(...........)))))(((((....-----.(((((...(......)...)))))..)))))......)))))).)))... ( -30.40) >DroAna_CAF1 89422 80 - 1 CCCGGUGCCGAUUGG--CA----------GUCGAAC--G--GAGA---GCUGCGAUGCCCGACCGAAAUGCUGAAUAAGCGGCAAGUGGCCCGCGGAGG (((((.((((.(((.--(.----------.(((..(--(--(...---((......)))))..)))...(((.....)))).))).))))))).))... ( -27.60) >consensus CCCGGUGCCAAUUGG__CAAGUUACAAUAGCCGAGCUUGUCGGGG___GCCGCGAGGUGGGUGGAAAAUGCUGAAUGAGCGGAAAGUGGCACAGGGAAA (((.((((((...................(((...((((.(((......)))))))...)))......((((.....)))).....)))))).)))... (-18.60 = -19.93 + 1.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:24 2006