
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
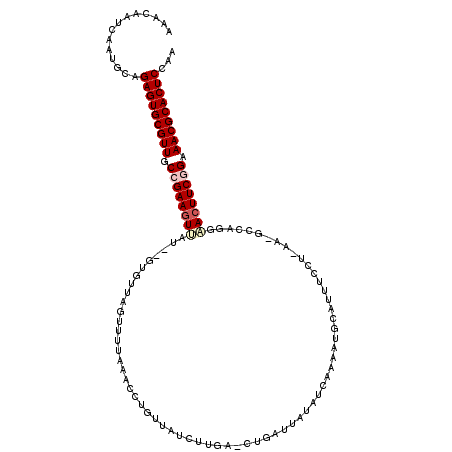
| Location | 2,548,923 – 2,549,040 |
| Length | 117 |
| Max. P | 0.998952 |

| Location | 2,548,923 – 2,549,040 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -19.65 |
| Energy contribution | -20.49 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
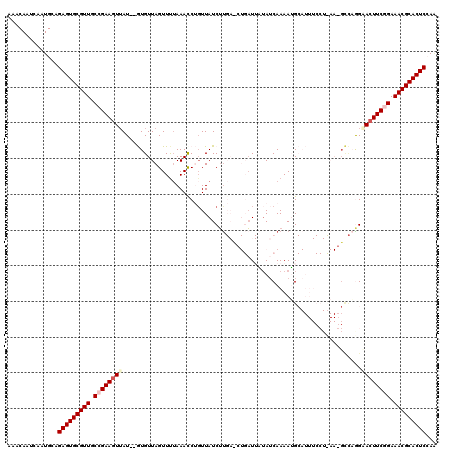
>3L_DroMel_CAF1 2548923 117 + 23771897 AAACAAUCAAUGCAGAGUGCGUUGCCGAAGUUAU--GUGUUAGUUUUAAAUCUGUUAUCAUGAUCUGUUUA-AUCAAAAUGCAUUUCCUCAACGCCCGGCACUUCGGAAACGCACUCCAA ..............(((((((((.(((((((.((--(((((....((((((..(((.....)))..)))))-)....))))))).........((...))))))))).)))))))))... ( -31.20) >DroSec_CAF1 10931 118 + 1 AAACAAUCAAUGCAGAGUGCGUUGCCGAAGUUAU--GUGUUAGUUUUAAACCUGUUAUCUUGACCUGAUUAUAUCAAAAUGCAUUUCCUUAAGGCCAAGUACUUCGGAAACGCACUCAAA ..............(((((((((.(((((((...--(((.(((........))).)))((((.(((.........................))).)))).))))))).)))))))))... ( -30.71) >DroSim_CAF1 7835 118 + 1 AAACAAUCAAUGCAGAGUGCGUUGCCGAAAUAAU--GUGUUAGUUUUAAACCUCUUAUCUUGACCUGAUUAUAUCAAAAUGCAUUUCCUUAAGGCCAAGUACUUCGGAAACGCACUCAAG ..............(((((((((.(((((.....--..(((.......))).......((((.(((.........................))).))))...))))).)))))))))... ( -26.51) >DroEre_CAF1 24667 96 + 1 AAACAAUCAAUGAAGAGUGCGUUCCUGAAGUUAU--AUGUUAGUUCGAAACCAGUUAUCUC------AU-UUAUCAAAAGG---------------CGGAACUUCUGAAACGCACUCCAU ..............(((((((((.(.((((((..--.((((..((..(((..((....)).------.)-))...))..))---------------)).)))))).).)))))))))... ( -22.30) >DroYak_CAF1 8959 101 + 1 AAACAAUCAAUGCAGAGUGCGUUGCCGAAGUUAUAUAUGUUAGUUUUAAACCAGUUAUCUU------AU-UUAACAAAAAG------------GUUAGGAACUUCUGAAACGCACUCCAA ..............(((((((((.(.((((((....((.((..(.(((((..((....)).------.)-)))).)..)).------------))....)))))).).)))))))))... ( -22.80) >consensus AAACAAUCAAUGCAGAGUGCGUUGCCGAAGUUAU__GUGUUAGUUUUAAACCUGUUAUCUUGA_CUGAUUAUAUCAAAAUGCAUUUCCU_AA_GCCAGGAACUUCGGAAACGCACUCCAA ..............(((((((((.((((((((...................................................................)))))))).)))))))))... (-19.65 = -20.49 + 0.84)

| Location | 2,548,923 – 2,549,040 |
|---|---|
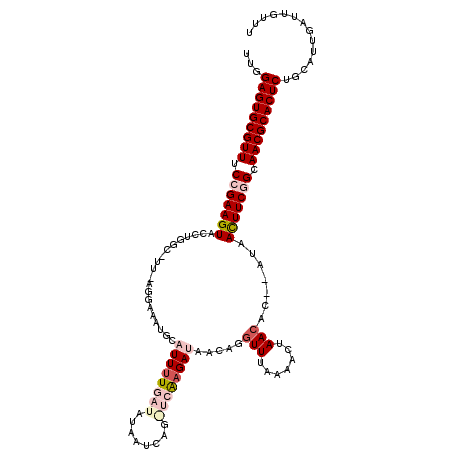
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -22.28 |
| Energy contribution | -24.28 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
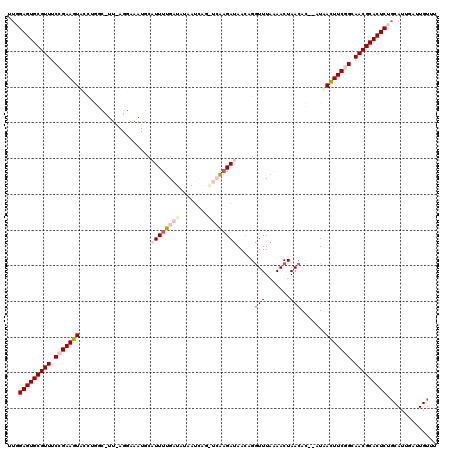
>3L_DroMel_CAF1 2548923 117 - 23771897 UUGGAGUGCGUUUCCGAAGUGCCGGGCGUUGAGGAAAUGCAUUUUGAU-UAAACAGAUCAUGAUAACAGAUUUAAAACUAACAC--AUAACUUCGGCAACGCACUCUGCAUUGAUUGUUU ..((((((((((.(((((((....((((((.....)))))(((.((((-(.....))))).)))...................)--...))))))).))))))))))............. ( -36.00) >DroSec_CAF1 10931 118 - 1 UUUGAGUGCGUUUCCGAAGUACUUGGCCUUAAGGAAAUGCAUUUUGAUAUAAUCAGGUCAAGAUAACAGGUUUAAAACUAACAC--AUAACUUCGGCAACGCACUCUGCAUUGAUUGUUU ...(((((((((.(((((((.((((((((....((((....))))(((...))))))))))).......(((.......)))..--...))))))).))))))))).............. ( -35.50) >DroSim_CAF1 7835 118 - 1 CUUGAGUGCGUUUCCGAAGUACUUGGCCUUAAGGAAAUGCAUUUUGAUAUAAUCAGGUCAAGAUAAGAGGUUUAAAACUAACAC--AUUAUUUCGGCAACGCACUCUGCAUUGAUUGUUU ...(((((((((.((((((((((((((((....((((....))))(((...))))))))))).......(((.......)))..--..)))))))).))))))))).............. ( -34.20) >DroEre_CAF1 24667 96 - 1 AUGGAGUGCGUUUCAGAAGUUCCG---------------CCUUUUGAUAA-AU------GAGAUAACUGGUUUCGAACUAACAU--AUAACUUCAGGAACGCACUCUUCAUUGAUUGUUU ..((((((((((((.((((((...---------------...(((((..(-((------.((....)).)))))))).......--..)))))).))))))))))))............. ( -26.46) >DroYak_CAF1 8959 101 - 1 UUGGAGUGCGUUUCAGAAGUUCCUAAC------------CUUUUUGUUAA-AU------AAGAUAACUGGUUUAAAACUAACAUAUAUAACUUCGGCAACGCACUCUGCAUUGAUUGUUU ..((((((((((.(.((((((......------------.......((((-((------.((....)).)))))).............)))))).).))))))))))............. ( -24.67) >consensus UUGGAGUGCGUUUCCGAAGUACCUGGC_UU_AGGAAAUGCAUUUUGAUAUAAUCAG_UCAAGAUAACAGGUUUAAAACUAACAC__AUAACUUCGGCAACGCACUCUGCAUUGAUUGUUU ...(((((((((.(((((((....................((((((((........)))))))).....(((.......))).......))))))).))))))))).............. (-22.28 = -24.28 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:28 2006