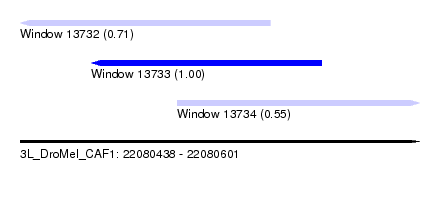
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,080,438 – 22,080,601 |
| Length | 163 |
| Max. P | 0.997881 |

| Location | 22,080,438 – 22,080,540 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -26.29 |
| Energy contribution | -27.23 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22080438 102 - 23771897 AUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAUAAAAUGGGCACAACA-----CCAGCGUCGUCGAA ........((.((((((...........)))))).)).((((.(((.((((((((((((....................))).)))))-----)))).))).)))). ( -39.85) >DroSec_CAF1 65869 102 - 1 AUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAUAAAAUGGGCACAACA-----CCAGCGUCGUCGAA ........((.((((((...........)))))).)).((((.(((.((((((((((((....................))).)))))-----)))).))).)))). ( -39.85) >DroSim_CAF1 72222 102 - 1 AUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAUAAAAUGGGCACAACA-----CCAGCGUCGUCGAA ........((.((((((...........)))))).)).((((.(((.((((((((((((....................))).)))))-----)))).))).)))). ( -39.85) >DroEre_CAF1 66054 107 - 1 AUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAUAAAAUGGGCACAACACCACACCAGCGUCGUCGGA ........((.((((((...........)))))).)).((((.(((.(((((((((((...)))))...........(((........))))))))).))).)))). ( -36.90) >DroYak_CAF1 80946 102 - 1 AUAAAACGACCGCUGCAAAAUGCCAGAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAAGCUAAGAACAAUAAAAUGGCCACAGCA-----CCAGCGUCGUCGGA ........((.((((((...........)))))).)).((((.(((.(((((((((((((.................))))))..)))-----)))).))).)))). ( -39.53) >DroAna_CAF1 87577 101 - 1 AUA-AACGGGCGCUGAAAAAUGCCAAAAUGCAGCCGAGGCGAGGACCCUGGUGAUGGCCGGGCCAAGAACAAUAAAAUGUGCACAGCA-----CCAGCGUCGAAAUA ...-....(((((((.....(((......)))(((..(((.(..((....))..).))).)))...............((((...)))-----))))))))...... ( -29.60) >consensus AUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAUAAAAUGGGCACAACA_____CCAGCGUCGUCGAA ........((.((((((...........)))))).)).((((.(((.((((((((((((....................))).))))).....)))).))).)))). (-26.29 = -27.23 + 0.95)


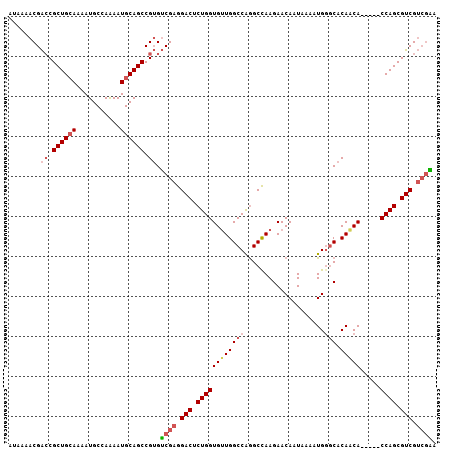
| Location | 22,080,467 – 22,080,561 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -20.86 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22080467 94 - 23771897 CAAAAUAAAAAAGAUACGAAAAUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAU ...........................(((((((((((...........))))).)).)))).((.(.(((((....))))))))......... ( -22.50) >DroSec_CAF1 65898 94 - 1 CAAAAUAAAAAAGAUACGAAAAUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAU ...........................(((((((((((...........))))).)).)))).((.(.(((((....))))))))......... ( -22.50) >DroSim_CAF1 72251 94 - 1 CAAAAUAAAAAAGAUACGAAAAUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAU ...........................(((((((((((...........))))).)).)))).((.(.(((((....))))))))......... ( -22.50) >DroEre_CAF1 66088 94 - 1 CAAAAUAAAAAAGAUACGAAAAUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAU ...........................(((((((((((...........))))).)).)))).((.(.(((((....))))))))......... ( -22.50) >DroYak_CAF1 80975 94 - 1 CAAAAUAAAAAAGAUACGAAAAUAAAACGACCGCUGCAAAAUGCCAGAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAAGCUAAGAACAAU ...........................(((((((((((...........))))).)).)))).((.((.((((....))))))))......... ( -17.50) >consensus CAAAAUAAAAAAGAUACGAAAAUAAAACGACCGCUGCAAAAUGCCAAAAUGCAGCCGUGUCGAGGACUCUGGUGUUGGCCAGGCCAAGAACAAU ...........................(((((((((((...........))))).)).)))).((.(.(((((....))))))))......... (-20.86 = -20.90 + 0.04)

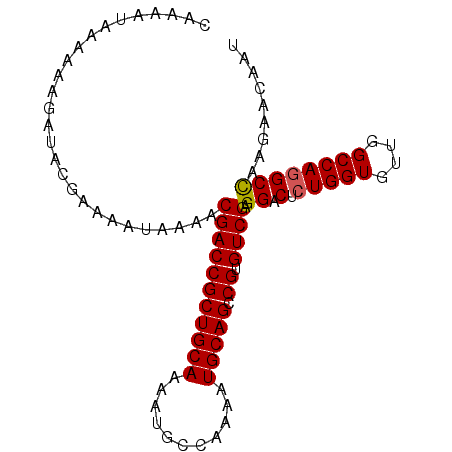
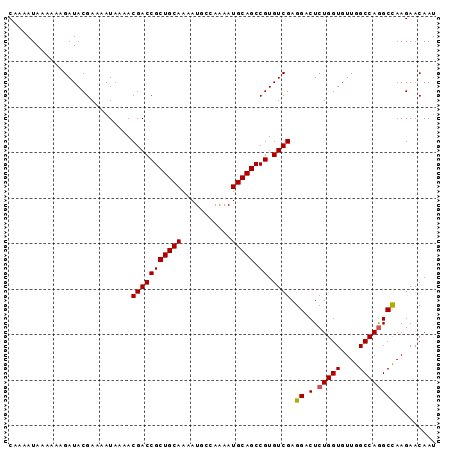
| Location | 22,080,502 – 22,080,601 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.63 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22080502 99 + 23771897 CACGGCUGCAUUUUGGCAUUUUGCAGCGGUCGUUUUAUUUUCGUAUCUUUUUUAUUUUGAAGUUUAAUGCCAGAGUCGUGCCUCAUAUUUCGACGAUGG ...(((.(((((((((((((..((.((((...........)))).((...........)).))..))))))))))).)))))................. ( -21.90) >DroSec_CAF1 65933 99 + 1 CACGGCUGCAUUUUGGCAUUUUGCAGCGGUCGUUUUAUUUUCGUAUCUUUUUUAUUUUGAAGUUUAAUGCCAGAGUCGUGCCUCAUAUUUCGACGAUGG ...(((.(((((((((((((..((.((((...........)))).((...........)).))..))))))))))).)))))................. ( -21.90) >DroSim_CAF1 72286 99 + 1 CACGGCUGCAUUUUGGCAUUUUGCAGCGGUCGUUUUAUUUUCGUAUCUUUUUUAUUUUGAAGUUUAAUGCCAGAGUCGUGCCUCAUAUUUCGACGAUGG ...(((.(((((((((((((..((.((((...........)))).((...........)).))..))))))))))).)))))................. ( -21.90) >DroEre_CAF1 66123 99 + 1 CACGGCUGCAUUUUGGCAUUUUGCAGCGGUCGUUUUAUUUUCGUAUCUUUUUUAUUUUGAAGUUUAAUGCCAGAGUCGUGCCUCAUAUUUCGACGAUGG ...(((.(((((((((((((..((.((((...........)))).((...........)).))..))))))))))).)))))................. ( -21.90) >DroYak_CAF1 81010 99 + 1 CACGGCUGCAUUCUGGCAUUUUGCAGCGGUCGUUUUAUUUUCGUAUCUUUUUUAUUUUGAAGUUUAAUGCCAGAGUCGUGCCUCAUAUUUCGACGAUGG ...(((.(((((((((((((..((.((((...........)))).((...........)).))..))))))))))).)))))................. ( -24.50) >DroAna_CAF1 87641 96 + 1 CUCGGCUGCAUUUUGGCAUUUUUCAGCGCCCGUU-UAUUUUUGUG--UUUUUUAGUUUGAAGUUUAAUGCCAGAGUCGUGCCUCAUAUUUCGACGAUGG ...(((.((((((((((((((((((((.......-(((....)))--.......).))))))...))))))))))).)))))................. ( -20.34) >consensus CACGGCUGCAUUUUGGCAUUUUGCAGCGGUCGUUUUAUUUUCGUAUCUUUUUUAUUUUGAAGUUUAAUGCCAGAGUCGUGCCUCAUAUUUCGACGAUGG ...(((.(((((((((((((..((.((((...........)))).....(((......)))))..))))))))))).)))))................. (-20.58 = -20.63 + 0.06)

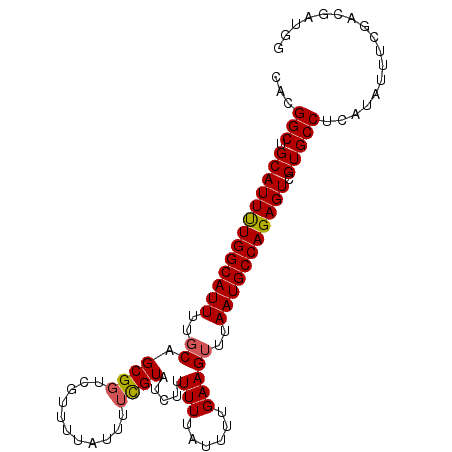
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:21 2006