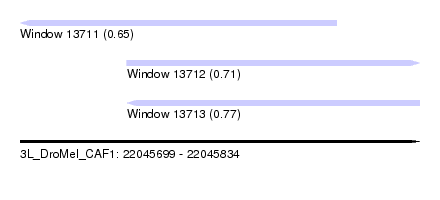
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,045,699 – 22,045,834 |
| Length | 135 |
| Max. P | 0.771190 |

| Location | 22,045,699 – 22,045,806 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -15.39 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22045699 107 - 23771897 UUGGUUUGGCAAUUUUGUAAGCGAAAU-UGCCGCUGUCUCGUUUUUCUGCGUUUUCAUUUUCGUCG-CUGGCU--------UU---UUUUGUGCCGCGCAGUUAUUAGAAGAAAAUGCUU .......((((((((((....))))))-))))................((((((((.((((.((.(-(((((.--------..---.........)).)))).)).)))))))))))).. ( -29.50) >DroPse_CAF1 43390 116 - 1 UUGGUUUGGCAAUUCUGUAAGCGAAAU-UGCCGCUGUCUCGUUUUUCUGCGUUUUCAUUUUCAUCGUCUCGUUUUUCUUCUUU---UUUUAUCCUGCGCAGUUAUUAGAAGAAAAUGCCU ..(((..(((((((.((....)).)))-))))....((((......((((((...............................---.........))))))......).)))....))). ( -19.90) >DroGri_CAF1 30994 82 - 1 UUGGUUUGGCAAUUUUGUAAGCGAAAU-UGCCACUGUCUCGCAUU----CGUUGUCGUUUU--------------------------------UUGCGCAGUUAUUAGAAGAA-AUGCCG ..(((.(((((((((((....))))))-)))))(((.((((((..----((....))....--------------------------------.)))).))....))).....-..))). ( -22.90) >DroEre_CAF1 31370 107 - 1 UUGGUUUGGCAAUUUUGUAAGCGAAAU-UGCCGCUGUCUCGUUUUUCUGCGUUUUCAUUUUCGUUG-CUGGCU--------UU---UUUUCUGCCGCGCAGUUAUUAGAAGAAAAUGCUU .......((((((((((....))))))-))))................((((((((.(((...(((-(((((.--------..---......)))).)))).....))).)))))))).. ( -31.00) >DroAna_CAF1 46394 106 - 1 UUGGUUUGGCAAUUUUGUAAGCGAAAU-UGCCGCUGUCUCGUUUUUCUGCGUUUUCAUUUUCAUCG-CUCGCU--------UU----UUUCUGCUGCGCAGUUAUUAGAAGAAAAUGUUU .......((((((((((....))))))-))))................((((((((.((((.((.(-(((((.--------..----........))).))).)).)))))))))))).. ( -27.00) >DroPer_CAF1 43194 120 - 1 UUGGUUUGGCAAUUCUGUAAGCGAAAUUUGCCGCUGUCUCGUUUUUCUGCGUUUUCAUUUUCAUCGUCUCGUUUUUCUUCUUUUUUUUUUAUCCUGCGCAGUUAUUAGAAGAAAAUGCCU .......((((.((((...((((........)))).(((.((....((((((...........................................))))))..)).)))))))..)))). ( -19.33) >consensus UUGGUUUGGCAAUUUUGUAAGCGAAAU_UGCCGCUGUCUCGUUUUUCUGCGUUUUCAUUUUCAUCG_CUCGCU________UU___UUUUAUGCUGCGCAGUUAUUAGAAGAAAAUGCCU .......((((((((((....)))))).))))((..((((......((((((...........................................))))))......).)))....)).. (-15.39 = -16.28 + 0.89)



| Location | 22,045,735 – 22,045,834 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -15.75 |
| Consensus MFE | -11.72 |
| Energy contribution | -11.58 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22045735 99 + 23771897 AGCCAGCGACGAAAAUGAAAACGCAGAAAAACGAGACAGCGGCAAUUUCGCUUACAAAAUUGCCAAACCAAAAAGGACUUUGAAUUCCAAUUGCAAAG-----------A ..((.(((.............)))................((((((((........))))))))..........)).((((((((....))).)))))-----------. ( -16.02) >DroVir_CAF1 37641 84 + 1 -----------AAAACGAUAACG----AGUGCGAGACAGCGGCAAUUUCGCUUACAAAAUUGCCAAACCAAAAAUGACUUUGAAUUCCAAUUGCAAAG-----------A -----------..........(.----..(((((......((((((((........))))))))....((((......))))........)))))..)-----------. ( -14.60) >DroGri_CAF1 31020 84 + 1 -----------AAAACGACAACG----AAUGCGAGACAGUGGCAAUUUCGCUUACAAAAUUGCCAAACCAAAAAUGACUUUGAAUUCCAAUUGCGCCG-----------A -----------..........((----..(((((.....(((((((((........)))))))))...((((......))))........))))).))-----------. ( -14.60) >DroWil_CAF1 44865 82 + 1 ----------------AAAAACGCACA-AAACGAGACAGCGGCAAUUUCGCUUACAAAAUUGCCAAACCAAAAAGGACUUUGAAUUCCAAUUGCAAAG-----------C ----------------......(((..-............((((((((........))))))))..........(((........)))...)))....-----------. ( -14.50) >DroMoj_CAF1 41180 84 + 1 -----------AAAACGAUAACG----AGUGCGAGACAGCGGCAAUUUCGCUUACAAAAUUGCCAAACCAAAAAUGACUUUGAAUUCCAAUUGCGAUG-----------A -----------............----..(((((......((((((((........))))))))....((((......))))........)))))...-----------. ( -13.60) >DroAna_CAF1 46429 110 + 1 AGCGAGCGAUGAAAAUGAAAACGCAGAAAAACGAGACAGCGGCAAUUUCGCUUACAAAAUUGCCAAACCAAAAAGGACUUUGAAUUCCAAUUGCAAAGUUGGAAAGUACA .(((..(.((....)))....))).......((((.....((((((((........))))))))...((.....))..))))..((((((((....))))))))...... ( -21.20) >consensus ___________AAAACGAAAACG____AAAACGAGACAGCGGCAAUUUCGCUUACAAAAUUGCCAAACCAAAAAGGACUUUGAAUUCCAAUUGCAAAG___________A ......................................((((((((((........))))))))....((((......))))..........))................ (-11.72 = -11.58 + -0.14)

| Location | 22,045,735 – 22,045,834 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -19.94 |
| Consensus MFE | -17.00 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22045735 99 - 23771897 U-----------CUUUGCAAUUGGAAUUCAAAGUCCUUUUUGGUUUGGCAAUUUUGUAAGCGAAAUUGCCGCUGUCUCGUUUUUCUGCGUUUUCAUUUUCGUCGCUGGCU .-----------....((....((((.((((((....))))))...((((((((((....))))))))))............))))(((.............)))..)). ( -20.32) >DroVir_CAF1 37641 84 - 1 U-----------CUUUGCAAUUGGAAUUCAAAGUCAUUUUUGGUUUGGCAAUUUUGUAAGCGAAAUUGCCGCUGUCUCGCACU----CGUUAUCGUUUU----------- .-----------...(((.....((((.(((((....)))))))))((((((((((....))))))))))........)))..----............----------- ( -18.60) >DroGri_CAF1 31020 84 - 1 U-----------CGGCGCAAUUGGAAUUCAAAGUCAUUUUUGGUUUGGCAAUUUUGUAAGCGAAAUUGCCACUGUCUCGCAUU----CGUUGUCGUUUU----------- .-----------.((((((((.(((..((((((....))))))..(((((((((((....)))))))))))..........))----))))).))))..----------- ( -21.70) >DroWil_CAF1 44865 82 - 1 G-----------CUUUGCAAUUGGAAUUCAAAGUCCUUUUUGGUUUGGCAAUUUUGUAAGCGAAAUUGCCGCUGUCUCGUUU-UGUGCGUUUUU---------------- (-----------(....(((..(((........)))...)))....((((((((((....)))))))))))).....(((..-...))).....---------------- ( -19.20) >DroMoj_CAF1 41180 84 - 1 U-----------CAUCGCAAUUGGAAUUCAAAGUCAUUUUUGGUUUGGCAAUUUUGUAAGCGAAAUUGCCGCUGUCUCGCACU----CGUUAUCGUUUU----------- .-----------....((.....((((.(((((....)))))))))((((((((((....))))))))))........))...----............----------- ( -18.30) >DroAna_CAF1 46429 110 - 1 UGUACUUUCCAACUUUGCAAUUGGAAUUCAAAGUCCUUUUUGGUUUGGCAAUUUUGUAAGCGAAAUUGCCGCUGUCUCGUUUUUCUGCGUUUUCAUUUUCAUCGCUCGCU ......((((((........)))))).((((((....))))))...((((((((((....))))))))))................(((.............)))..... ( -21.52) >consensus U___________CUUUGCAAUUGGAAUUCAAAGUCAUUUUUGGUUUGGCAAUUUUGUAAGCGAAAUUGCCGCUGUCUCGCAUU____CGUUUUCGUUUU___________ ................((.........((((((....))))))..(((((((((((....))))))))))).......)).............................. (-17.00 = -16.62 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:14:01 2006