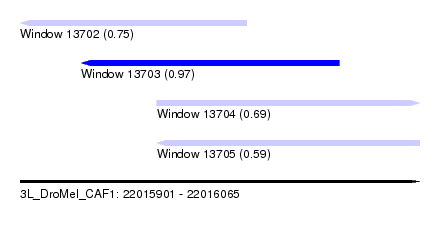
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,015,901 – 22,016,065 |
| Length | 164 |
| Max. P | 0.974679 |

| Location | 22,015,901 – 22,015,994 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.07 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
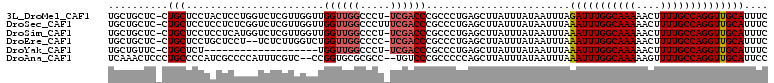
>3L_DroMel_CAF1 22015901 93 - 23771897 UUUAGAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUC---------CAACCAUCCGAGUGGGUGAACUUUACUCUUAUGCAAAUGACA---CACUCACACACACA---- .(..(((.(((((((....)))))))(((((......---------))))))))..)(((((((........................---))))))).......---- ( -21.56) >DroVir_CAF1 12526 99 - 1 UUUAAAUUUGGCAAAAG-U-UUG-CAGGUUGCAUUCAAAUUUUCCGCUGCCGU-CGAGUGGGUGAACUUUACUUUUAUGCAAAUGACACUGC------AAACACUAACA ................(-(-(((-(((((.((.............)).)))((-((..((.(((((.......))))).))..))))..)))------))))....... ( -22.52) >DroPse_CAF1 18552 90 - 1 UUUAAAUUUGGCAAAAAGUUUUGCCAGGUUGCAUUCC---------CAGCCAU-CGAGUGGGUGAACUUUACUUUUAUGUAAAUGCCG---C------ACACACUAUCA ........(((((((....)))))))(((((......---------)))))..-..((((.(((..((((((......))))).)...---)------)).)))).... ( -23.50) >DroGri_CAF1 8849 100 - 1 UUUAAAUUUGGCAAAAG-UUUUG-CAGGUUGCAUUCAAAUUUUCCGCUGCCGU-CGAGUGGGCGAACUUUACUUUUAUGCAAAUGACACUGC------AAACACUAACA ...............((-(((((-(((.((((((.............((((..-......))))............))))))......))))------))).))).... ( -19.21) >DroAna_CAF1 8306 87 - 1 UUUAAAUUUGGCAAAAAGUUUUGCCAGGUUGCAUUCC---------CAGCCAUCCGAGUGGGUGAACUUUACUUUUAUGCAAAUGCCA---U------ACACAGU---- ....(((((((((((....)))))))))))(((((..---------..((.((..(((((((....).))))))..)))).)))))..---.------.......---- ( -19.10) >DroPer_CAF1 18552 90 - 1 UUUAAAUUUGGCAAAAAGUUUUGCCAGGUUGCAUUCC---------CAGCCAU-CGAGUGGGUGAACUUUACUUUUAUGUAAAUGCCG---C------ACACACUAUCA ........(((((((....)))))))(((((......---------)))))..-..((((.(((..((((((......))))).)...---)------)).)))).... ( -23.50) >consensus UUUAAAUUUGGCAAAAAGUUUUGCCAGGUUGCAUUCC_________CAGCCAU_CGAGUGGGUGAACUUUACUUUUAUGCAAAUGACA___C______ACACACUA_CA ....(((((((((((....)))))))))))((((............((.(((......))).))............))))............................. (-14.81 = -15.07 + 0.25)
| Location | 22,015,926 – 22,016,032 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -24.20 |
| Energy contribution | -23.73 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22015926 106 - 23771897 UUGGUUGGCCCUUCGACCCGCCCUGAGCUUAUUUAUAAUUUAGAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUCCAACCAUCCGAGUGGGUGAACUUUACUCUUA .(((((((.....(((((....(((((.(((....))))))))...(((((((....)))))))))))).....)))))))...(((((((....).))))))... ( -30.90) >DroPse_CAF1 18575 95 - 1 -------GCCCU---CUCCACCCUCAGCUUAUUUAUAAUUUAAAUUUGGCAAAAAGUUUUGCCAGGUUGCAUUCCCAGCCAU-CGAGUGGGUGAACUUUACUUUUA -------((((.---(((............................(((((((....)))))))(((((......)))))..-.))).)))).............. ( -24.20) >DroEre_CAF1 7642 106 - 1 CUGGUUGGCCCCUCGACCCGCCCUGAGCUUAUUUAUAAUUUAAAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUCCAACCAUCCGAGUGGGUGAACUUUACUUUUA ..((((.((((((((...............................(((((((....)))))))(((((......)))))...)))).)))).))))......... ( -31.30) >DroYak_CAF1 8577 105 - 1 -UGGUUGGCCCUUCGACCCGCCCUGAGCUUAUUUAUAAUUUAAAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUCCAACCAUUCGAGUGGGUGAACUUUACUUUUA -.((((.(((((((((..............................(((((((....)))))))(((((......)))))..))))).)))).))))......... ( -29.80) >DroAna_CAF1 8325 105 - 1 CCGGUGCGCGCC-UGUCCCGCCCCCAGCUUAUUUAUAAUUUAAAUUUGGCAAAAAGUUUUGCCAGGUUGCAUUCCCAGCCAUCCGAGUGGGUGAACUUUACUUUUA ..(((....)))-.....(((((...((((................(((((((....)))))))(((((......)))))....)))))))))............. ( -26.50) >DroPer_CAF1 18575 95 - 1 -------GCCCU---CUCCACCCUCAGCUUAUUUAUAAUUUAAAUUUGGCAAAAAGUUUUGCCAGGUUGCAUUCCCAGCCAU-CGAGUGGGUGAACUUUACUUUUA -------((((.---(((............................(((((((....)))))))(((((......)))))..-.))).)))).............. ( -24.20) >consensus _UGGUUGGCCCU_CGACCCGCCCUCAGCUUAUUUAUAAUUUAAAUUUGGCAAAAACUUUUGCCAGGUUGCAUUCCCAACCAUCCGAGUGGGUGAACUUUACUUUUA ..................(((((...((((................(((((((....)))))))(((((......)))))....)))))))))............. (-24.20 = -23.73 + -0.47)

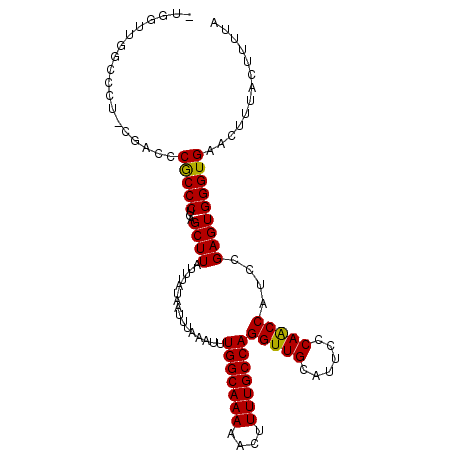
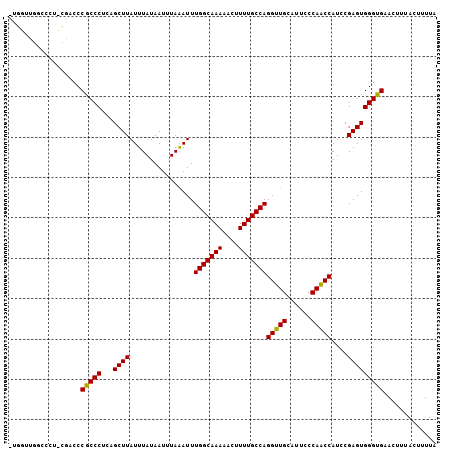
| Location | 22,015,957 – 22,016,065 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -18.46 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22015957 108 + 23771897 GAAAUGCAACCUGGCAAAAGUUUUUGCCAAAUCUAAAUUAUAAAUAAGCUCAGGGCGGGUCGA-AGGGCCAACCAACCAACGAGACCAGGAGUAGGAGCAG-GAGCAGCA ....(((....(((((((....)))))))..................((((..((..((((..-..))))..))..((.((..........)).)).....-)))).))) ( -30.50) >DroSec_CAF1 6914 109 + 1 GAAAUGCAACCUGGCAAAAGUUUUUGCCAAAUUUAAAUUAUAAAUAAGCUCAGGGCGGGUCGAAAGGGCCAACCAACCAACGAGACCGAGAGGAGGAGCAG-GAGCAGCA ....(((..(((((((((....))))))...................((((.((..((.((....)).))..))...........((....))..))))))-).)))... ( -32.80) >DroSim_CAF1 7282 108 + 1 GAAAUGCAACCUGGCAAAAGUUUUUGCCAAAUUUAAAUUAUAAAUAAGCUCAGGGCGGGUCGA-AGGGCCAACCAACCAACGAGACCAUGAGGAGGAGCAG-GAGCAGCA ....(((..(((((((((....))))))...................((((..((..((((..-..))))..))...........((....))..))))))-).)))... ( -31.10) >DroEre_CAF1 7673 106 + 1 GAAAUGCAACCUGGCAAAAGUUUUUGCCAAAUUUAAAUUAUAAAUAAGCUCAGGGCGGGUCGA-GGGGCCAACCAGACCAAGAGA--AGGAGCAGGAGCAG-GAGCAGCA ....(((..(((((((((....))))))...................((((....(.((((..-.((.....)).))))....).--..))))))).((..-..)).))) ( -29.40) >DroYak_CAF1 8608 89 + 1 GAAAUGCAACCUGGCAAAAGUUUUUGCCAAAUUUAAAUUAUAAAUAAGCUCAGGGCGGGUCGA-AGGGCCAACCA-------------------AGAGCAG-GAACAGCA ....(((..(((((((((....))))))...................((((..((..((((..-..))))..)).-------------------.))))))-)....))) ( -29.90) >DroAna_CAF1 8356 106 + 1 GGAAUGCAACCUGGCAAAACUUUUUGCCAAAUUUAAAUUAUAAAUAAGCUGGGGGCGGGACA--GGCGCGCACCGG--GACGAAAUGGGGCGAUGGGGCAGGGAGUUUGA ....(((......)))((((((((((((..(((....((((.......((.((.(((.(...--..).))).)).)--).....))))...)))..)))))))))))).. ( -28.30) >consensus GAAAUGCAACCUGGCAAAAGUUUUUGCCAAAUUUAAAUUAUAAAUAAGCUCAGGGCGGGUCGA_AGGGCCAACCAACCAACGAGACCAGGAGGAGGAGCAG_GAGCAGCA ....(((....(((((((....)))))))..................((((..((..((((.....))))..)).....................)))).....)))... (-18.46 = -19.18 + 0.72)

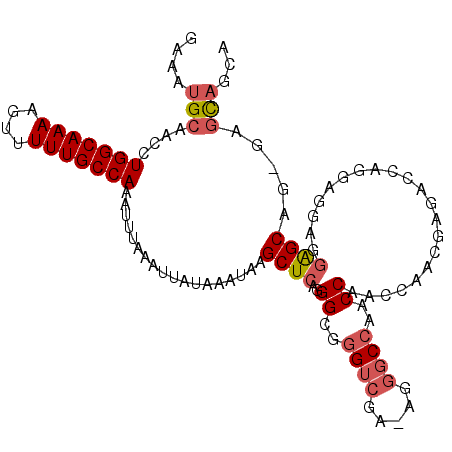
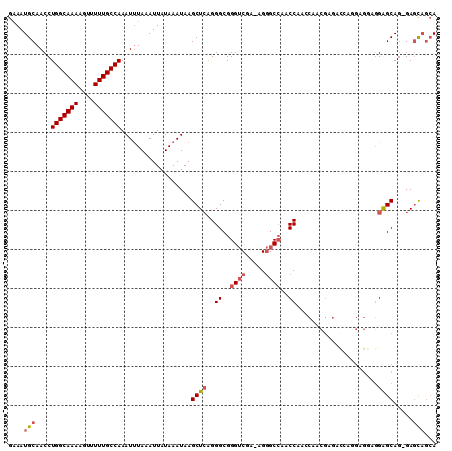
| Location | 22,015,957 – 22,016,065 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22015957 108 - 23771897 UGCUGCUC-CUGCUCCUACUCCUGGUCUCGUUGGUUGGUUGGCCCU-UCGACCCGCCCUGAGCUUAUUUAUAAUUUAGAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUC (((.((((-.....((.......)).......(((.((((((....-)))))).)))..))))..............(((((((((((....)))))))))))))).... ( -29.10) >DroSec_CAF1 6914 109 - 1 UGCUGCUC-CUGCUCCUCCUCUCGGUCUCGUUGGUUGGUUGGCCCUUUCGACCCGCCCUGAGCUUAUUUAUAAUUUAAAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUC ...(((..-..((((..((....)).......(((.((((((.....)))))).)))..))))..............(((((((((((....)))))))))))))).... ( -28.50) >DroSim_CAF1 7282 108 - 1 UGCUGCUC-CUGCUCCUCCUCAUGGUCUCGUUGGUUGGUUGGCCCU-UCGACCCGCCCUGAGCUUAUUUAUAAUUUAAAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUC ...(((..-..((((..((....)).......(((.((((((....-)))))).)))..))))..............(((((((((((....)))))))))))))).... ( -29.10) >DroEre_CAF1 7673 106 - 1 UGCUGCUC-CUGCUCCUGCUCCU--UCUCUUGGUCUGGUUGGCCCC-UCGACCCGCCCUGAGCUUAUUUAUAAUUUAAAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUC (((.((((-..((....))....--......((.(.((((((....-)))))).).)).))))..............(((((((((((....)))))))))))))).... ( -27.80) >DroYak_CAF1 8608 89 - 1 UGCUGUUC-CUGCUCU-------------------UGGUUGGCCCU-UCGACCCGCCCUGAGCUUAUUUAUAAUUUAAAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUC (((.....-..((((.-------------------.(((.((....-....)).)))..))))..............(((((((((((....)))))))))))))).... ( -25.60) >DroAna_CAF1 8356 106 - 1 UCAAACUCCCUGCCCCAUCGCCCCAUUUCGUC--CCGGUGCGCGCC--UGUCCCGCCCCCAGCUUAUUUAUAAUUUAAAUUUGGCAAAAAGUUUUGCCAGGUUGCAUUCC ..........(((...................--..((.(.(((..--.....))).))).................(((((((((((....)))))))))))))).... ( -18.10) >consensus UGCUGCUC_CUGCUCCUCCUCCUGGUCUCGUUGGUUGGUUGGCCCU_UCGACCCGCCCUGAGCUUAUUUAUAAUUUAAAUUUGGCAAAAACUUUUGCCAGGUUGCAUUUC ..........(((.......................((((((.....))))))........................(((((((((((....)))))))))))))).... (-16.99 = -17.52 + 0.53)
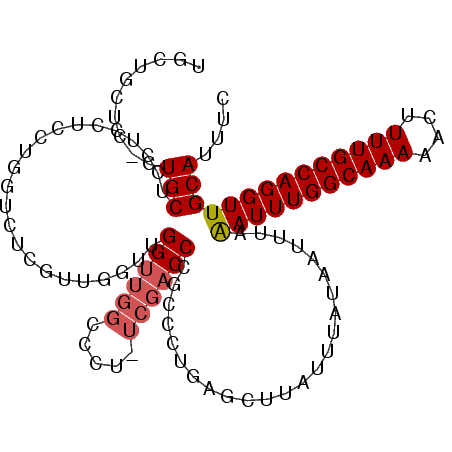
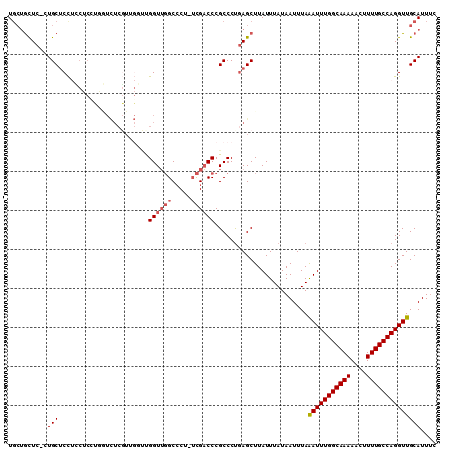
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:54 2006