
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,976,074 – 21,976,166 |
| Length | 92 |
| Max. P | 0.889626 |

| Location | 21,976,074 – 21,976,166 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -16.96 |
| Energy contribution | -18.48 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
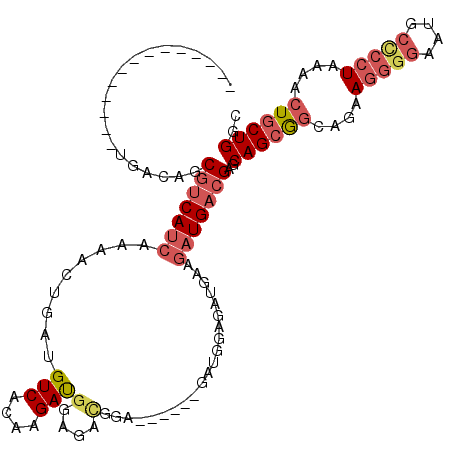
>3L_DroMel_CAF1 21976074 92 + 23771897 --------------UGACAGCGUCAUCAAAACUGAUGUCACAAGAUGGA------------GAUGGAGAUGAAGAUGACGAGCAGCAGCAGAAGGGGAAUGCCCCUAAAACUGCUGGC --------------......(((((((........((((.(......).------------))))........)))))))..((((((....(((((....)))))....)))))).. ( -28.39) >DroSec_CAF1 102704 98 + 1 --------------UGACAGCGUCAUCAAAACUGAUGUCACAAGAUGGAGACGGA------GAUGGAGAUGAAGAUGACGAGCAGCGGCAGAAGGGGAAUGCCCCUAAAACUGCUGGC --------------......(((((((.....(.((.((......((....))..------....)).)).).)))))))..((((((....(((((....)))))....)))))).. ( -30.80) >DroSim_CAF1 102952 98 + 1 --------------UGACAGCGUCAUCAAAACUGAUGUCACAAGAUGGAGACGGA------GAUGGAGAUGAAGAUGACGAGCAGCGGCAGAAGGGGAAUGCCCCUAAAACUGCUGGC --------------......(((((((.....(.((.((......((....))..------....)).)).).)))))))..((((((....(((((....)))))....)))))).. ( -30.80) >DroYak_CAF1 119484 104 + 1 --------------UGACAGCGUCAUCAAAACUGAUGUCACAAGAUGAAGAUGAAGAUGAAGAUGAAGAUGAAGAUGACGAGCAGCGGCAGAAGGAGAAUUCCCCUAAAACUGCUGGC --------------......(((((((....((...(((....)))..))((....))...............)))))))..((((((....(((.(....).)))....)))))).. ( -20.00) >DroAna_CAF1 106726 105 + 1 CACCGAACUUCCCUCGACGGCUCCACCAAAACUUAUGUCACACGACG---AUGUU------GAUGGAGAUG---AUGACGAGCAGGAGAAGAAAAGAAUUUCUCCUAAAA-UGCUGGC ..(((.......((((.((.(((((.(((......((((....))))---...))------).)))))...---.)).)))).((((((((.......))))))))....-...))). ( -25.50) >consensus ______________UGACAGCGUCAUCAAAACUGAUGUCACAAGAUGGAGACGGA______GAUGGAGAUGAAGAUGACGAGCAGCGGCAGAAGGGGAAUGCCCCUAAAACUGCUGGC ....................(((((((.........(((....)))(....).....................)))))))..((((((....(((((....)))))....)))))).. (-16.96 = -18.48 + 1.52)

| Location | 21,976,074 – 21,976,166 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21976074 92 - 23771897 GCCAGCAGUUUUAGGGGCAUUCCCCUUCUGCUGCUGCUCGUCAUCUUCAUCUCCAUC------------UCCAUCUUGUGACAUCAGUUUUGAUGACGCUGUCA-------------- (((((((((...(((((....)))))...)))))))...((((((..........((------------.(......).))..........)))))))).....-------------- ( -28.05) >DroSec_CAF1 102704 98 - 1 GCCAGCAGUUUUAGGGGCAUUCCCCUUCUGCCGCUGCUCGUCAUCUUCAUCUCCAUC------UCCGUCUCCAUCUUGUGACAUCAGUUUUGAUGACGCUGUCA-------------- ((.(((.((...(((((....)))))...)).)))))..(.((.(.(((((.....(------(..(((.(......).)))...))....))))).).)).).-------------- ( -23.90) >DroSim_CAF1 102952 98 - 1 GCCAGCAGUUUUAGGGGCAUUCCCCUUCUGCCGCUGCUCGUCAUCUUCAUCUCCAUC------UCCGUCUCCAUCUUGUGACAUCAGUUUUGAUGACGCUGUCA-------------- ((.(((.((...(((((....)))))...)).)))))..(.((.(.(((((.....(------(..(((.(......).)))...))....))))).).)).).-------------- ( -23.90) >DroYak_CAF1 119484 104 - 1 GCCAGCAGUUUUAGGGGAAUUCUCCUUCUGCCGCUGCUCGUCAUCUUCAUCUUCAUCUUCAUCUUCAUCUUCAUCUUGUGACAUCAGUUUUGAUGACGCUGUCA-------------- ((((((.((...(((((....)))))...)).))))...((((((.....((......((((...............))))....))....)))))))).....-------------- ( -21.86) >DroAna_CAF1 106726 105 - 1 GCCAGCA-UUUUAGGAGAAAUUCUUUUCUUCUCCUGCUCGUCAU---CAUCUCCAUC------AACAU---CGUCGUGUGACAUAAGUUUUGGUGGAGCCGUCGAGGGAAGUUCGGUG ((((((.-...((((((((.........))))))))((((.(..---...(((((((------((.((---.(((....)))....)).)))))))))..).))))....))).))). ( -30.20) >consensus GCCAGCAGUUUUAGGGGCAUUCCCCUUCUGCCGCUGCUCGUCAUCUUCAUCUCCAUC______UCCAUCUCCAUCUUGUGACAUCAGUUUUGAUGACGCUGUCA______________ ((.((((((...(((((....)))))......)))))).))....................................(((.((((......)))).)))................... (-17.18 = -17.42 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:39 2006