
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,953,290 – 21,953,565 |
| Length | 275 |
| Max. P | 0.893326 |

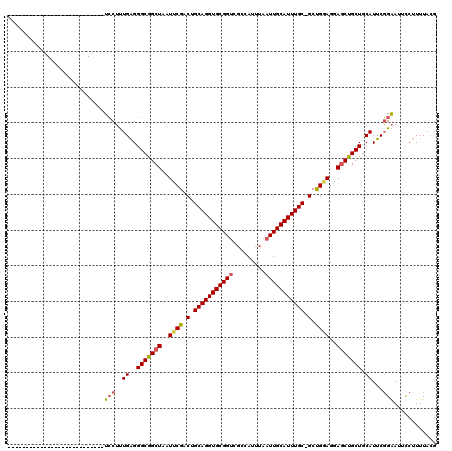
| Location | 21,953,290 – 21,953,382 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.08 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -28.41 |
| Energy contribution | -28.63 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
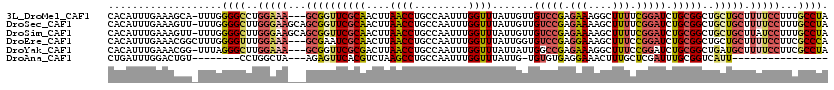
>3L_DroMel_CAF1 21953290 92 - 23771897 ---------------------------UCCUUUGAGGGCGGCUAAUUCGACUGCAGGUGCGGUCGCCAUUUAAUUGCAUUUGC-GCUGGAGGAGCUGCUGCAUUCGGAAUUCCUUUUACG ---------------------------(((..((..(((((((..((((..((((((((((((.........)))))))))))-).))))..))))))).))...)))............ ( -30.70) >DroSec_CAF1 79344 92 - 1 ---------------------------UCCUUUGAGGGCGGCUAAUUCGACUGCAGGUGCGGUCGCCAUUUAAUUGCAUUUGC-GCUGGAGGAGCUGCUGCAUUCGGAAUUCCUUUUACG ---------------------------(((..((..(((((((..((((..((((((((((((.........)))))))))))-).))))..))))))).))...)))............ ( -30.70) >DroSim_CAF1 78862 92 - 1 ---------------------------UCCUUUGAGGGCGGCUAAUUCGACUGCAGGUGCGGUCGCCAUUUAAUUGCAUUUGC-GCUGGAGGAGCUGCUGCAUUCGGAAUUCCUUUUACG ---------------------------(((..((..(((((((..((((..((((((((((((.........)))))))))))-).))))..))))))).))...)))............ ( -30.70) >DroEre_CAF1 75470 92 - 1 ---------------------------UCCUUUGAGGGCGGCUAAUUCGACUGCAGGUGCGGUCGCCAUUUAAUUGCAUUUGC-GCCGGAGGAGCUGCUGCAUUCGGAAUUCCUUUUACG ---------------------------(((..((..(((((((..((((..((((((((((((.........)))))))))))-).))))..))))))).))...)))............ ( -32.40) >DroYak_CAF1 90213 92 - 1 ---------------------------UCCUUUGAGGGCGGCUAAUUCGACUGCAGGUGCGGUCGCCAUUUAAUUGCAUUUGC-GCUGAAGGAGCUGCUGCAUUCGGAAUUCCUUUUAUG ---------------------------(((..((..(((((((..((((..((((((((((((.........)))))))))))-).))))..))))))).))...)))............ ( -31.20) >DroAna_CAF1 82396 120 - 1 GGAUUCGGAGGACGUCCUGACGUUCCAUGCACUGGGGGCAGCUAAUUCGGCAGCAGGUGCGGACUUUAUUUAAUUGCAUUUGCUGCCGCAGGAUCUGCUGCAUUCUGGAAUUCUUUUACG ((((((.(.((((((....))))))).......(((((((((..((((((((((((((((((...........)))))))))))))))...)))..))))).)))).))))))....... ( -45.60) >consensus ___________________________UCCUUUGAGGGCGGCUAAUUCGACUGCAGGUGCGGUCGCCAUUUAAUUGCAUUUGC_GCUGGAGGAGCUGCUGCAUUCGGAAUUCCUUUUACG ...........................(((..((..(((((((..((((.(.(((((((((((.........))))))))))).).))))..))))))).))...)))............ (-28.41 = -28.63 + 0.23)


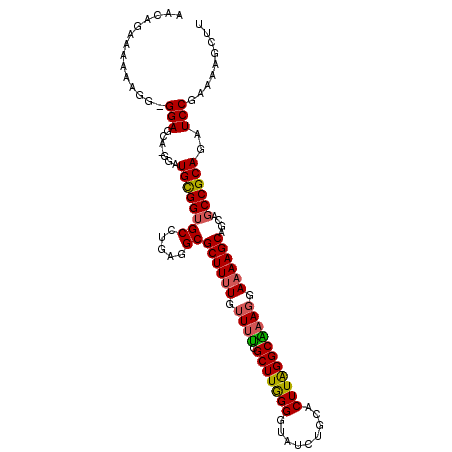
| Location | 21,953,382 – 21,953,489 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -29.46 |
| Energy contribution | -29.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21953382 107 + 23771897 AACAGAAUAAAGG-GGAGCA-AGAUGCGGAGCCUGAGGCGCUUUUGGUUUCGCUUAGGGUAUCUGCACUUAGGCAAAGGAAAAGCAGCAGCCGCAGAUCCGAAAAGCCU ...........((-(..((.-....))(((..(((.(((((((((..(((.(((((((((....)).))))))))))..))))))....))).))).)))......))) ( -31.20) >DroSec_CAF1 79436 108 + 1 AACAGAAAAAAGGGGGAGCA-GGAUGCGGUGCCUGAGGCGCUUUUGUUUUCGCUUGGGGUAUCUGCACUUAGGCAAAGGAAAAGCAGCAGCCGCAGAUCCGAAAAGCUU ...............((((.-((((((((((((...)))((((((.((((.(((((((((....)).))))))))))).))))))....))))))..))).....)))) ( -35.90) >DroSim_CAF1 78954 107 + 1 AACAGAAAAAAGG-GGAGCA-AGAUGCGGUGCCUGAGGCGCUUUUGUUUUCGCUUGGGGUAUCUGCACUUAGGCAAAGGAUAAGCAGCAGCCGCAGAUCCGAAAAGCUU .............-.(((((-((.((((((((((.(((((..........))))).))))).)))))))).......((((..((.......))..)))).....)))) ( -31.80) >DroEre_CAF1 75562 108 + 1 AACAGAAAAAAGG-GGAGCACGGAUGUGGUGCCUGGAGCGCUUUUGUUUCUGCUUGGGAUAUCUGCACUUGGGCGAAGGAAAAGCAGCAGCCGCAGAUCCGGAAAGCUU .............-.((((.((((((((((((.....))((((((.((((.(((..((.........))..))))))).))))))....))))))..))))....)))) ( -35.30) >DroYak_CAF1 90305 108 + 1 AACAGAAAAAAGA-GGAGCAAGGAUGCGGUGCCUAAGGCGCUUUUGUUGUUGCUUGGGGUAUCUGCACUUAGGCGAAGGAAAAGCAUCAGCCGCAGAUCCGGAAAGCCU ............(-((.....((((((((((((...)))((((((.((.(((((((((((....)).))))))))))).))))))....))))))..)))......))) ( -36.20) >consensus AACAGAAAAAAGG_GGAGCA_GGAUGCGGUGCCUGAGGCGCUUUUGUUUUCGCUUGGGGUAUCUGCACUUAGGCAAAGGAAAAGCAGCAGCCGCAGAUCCGAAAAGCUU ..............(((.......((((((((.....))((((((.((((.(((((((.........))))))))))).))))))....))))))..)))......... (-29.46 = -29.38 + -0.08)

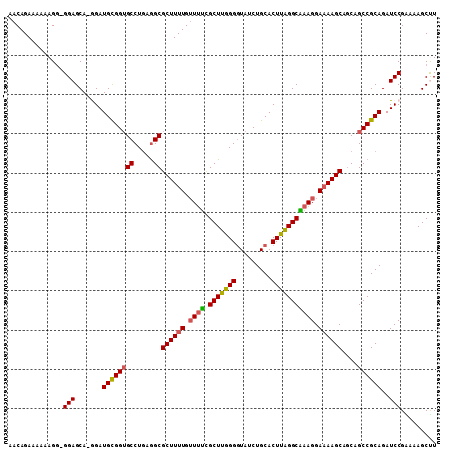
| Location | 21,953,449 – 21,953,565 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -18.63 |
| Energy contribution | -20.15 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21953449 116 - 23771897 CACAUUUGAAAGCA-UUUGGGGCCUGGAAA---GCGGUUCGCAACUUAACCUGCCAAUUUGGUUUAUUGUUGUCCGAGAAAGGCUUUUCGGAUCUGCGGCUGCUGCUUUUCCUUUGCCUA ..((..((....))-..))((((..(((((---((((((((((((..((((.........))))....)))(((((((((....)))))))))..))))..))))).)))))...)))). ( -37.40) >DroSec_CAF1 79504 119 - 1 CACAUUUGAAAGUU-UUUGGGGCUUGGGAAGCAGCGGUUCGCAACUUAACCUGCCAAUUUGGUUUAUUGUUGUCCGAGAAAAGCUUUUCGGAUCUGCGGCUGCUGCUUUUCCUUUGCCUA ..............-....((((..(((((((((((((.((((((..((((.........))))....)))(((((((((....)))))))))..))))))))))))..))))..)))). ( -45.50) >DroSim_CAF1 79021 119 - 1 CACAUUUGAAAGUU-UUUGGGGCUUGGGAAGCAGCGGUUCGCAACUUAACCUGCCAAUUUGGUUUAUUGUUGUCCGAGAAAAGCUUUUCGGAUCUGCGGCUGCUGCUUAUCCUUUGCCUA ..............-....((((..(((((((((((((.((((((..((((.........))))....)))(((((((((....)))))))))..))))))))))))..))))..)))). ( -45.50) >DroEre_CAF1 75630 117 - 1 CACAUUUGAAACGGCUUUGGGGUUUGGAAA---GCGAAUCGCAACUUAACCUGCCAAUUUGGUUUAUUGGUGUCCGAGGAAAGCUUUCCGGAUCUGCGGCUGCUGCUUUUCCUUCGCCCA ............(((...((((((((((((---((...(((..(((......(((.....))).....)))...))).....)))))))))))).((((...))))....))...))).. ( -33.60) >DroYak_CAF1 90373 116 - 1 CACAUUUGAAACGG-UUUAGGGCUUGGAAA---GCGGUUCGCGACUUAACCUGCCAAUUUGGUUUAUUAUUGGCCGAGAAAGGCUUUCCGGAUCUGCGGCUGAUGCUUUUCCUUCGCCUA ..............-....((((..(((((---((..(((.((.........((((((..........)))))))).)))..)))))))(((...(((.....)))...)))...)))). ( -32.50) >DroAna_CAF1 82538 92 - 1 CUGAUUUGGACUGU--------CCUGGCUA---AGAGUUCACGUCUAAGCCUGCCAAUUUGGUUUAUUG-UGUGUGAGGAAACUUUGCUCGAUUUGCGGUCAUU---------------- ........((((((--------...((((.---((((....).))).)))).(((.....)))..((((-.(((.(((....)))))).))))..))))))...---------------- ( -22.30) >consensus CACAUUUGAAAGGU_UUUGGGGCUUGGAAA___GCGGUUCGCAACUUAACCUGCCAAUUUGGUUUAUUGUUGUCCGAGAAAAGCUUUCCGGAUCUGCGGCUGCUGCUUUUCCUUUGCCUA ...................((((..(((((...((((((((((....((((.........)))).......(((((.(((....))).))))).)))))..))))).)))))...)))). (-18.63 = -20.15 + 1.52)
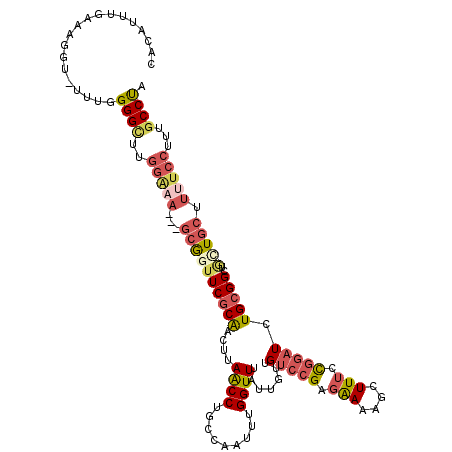
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:32 2006