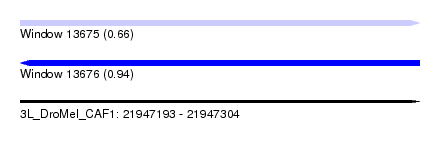
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,947,193 – 21,947,304 |
| Length | 111 |
| Max. P | 0.940644 |

| Location | 21,947,193 – 21,947,304 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.40 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.659710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21947193 111 + 23771897 UA---------GCCAACUCGAACGACCAACCGAAAGUAUUUAGUCACAAACUGUCACAUGUGAUUGUGACGCAUUCGGCUGGUCGGUUAACCCCAGCGGUUGUCAUUUUGCGGCCAGAGU ..---------....((((...((((((.(((((.((.....((((((....((((....)))))))))))).))))).))))))............((((((......)))))).)))) ( -37.80) >DroSec_CAF1 68867 111 + 1 UA---------GCCAACUCGAACGACCAACCGAAAGUAUUUAGUCACAAACUGUCACAUGUGAUUGUGACGCAUUCGGCUGGUCGGUUGGCCCCAGCAGUUGUCAUUUUGCGGCCAGAGU ..---------....((((...((((((.(((((.((.....((((((....((((....)))))))))))).))))).))))))..(((((...((((........))))))))))))) ( -42.00) >DroEre_CAF1 68648 119 + 1 UGGCCACAAUGGCCAACUCGAACGCCCAACGGAAAGUAUUUAGUCGCAAACUGUCACAUGUGAUUAUGACGCAUUCAGCUGGUCGGUCUACCC-AGCAGUUGUCAUUUUGUGGCCAAAGU (((((((((((((.((((.(((.((((...))...((...((((((((..........))))))))..)))).))).(((((.........))-))))))))))))..)))))))).... ( -39.20) >DroYak_CAF1 84719 111 + 1 UG---------ACCAGCUCAAAAGCGCAACCGAAAGUAUUUAGUCACAAACUGUCACAUGUGAUUGUGACGCAUUCAGCUGGUCGGUCUGCCCGAGCAGUUGUCAUUUUGUGCCCAGAGU ..---------....((((....((((((.(....)......((((((....((((....)))))))))).....((((((.((((.....)))).)))))).....))))))...)))) ( -34.80) >DroAna_CAF1 76983 110 + 1 UG---------CCCGACACGAAUUUGCAGCCAAAAGUAUUUAGUCACAAGCCGUCACAUGUGAUUGUGACGCAUUCGACUGGCCGGCUGUCCCCGGCAGUUGUCAU-UUGUGGCCAGAGU ..---------.....(((((((..(((((..........(((((....((.((((((......))))))))....)))))(((((......))))).))))).))-)))))........ ( -35.20) >consensus UG_________GCCAACUCGAACGACCAACCGAAAGUAUUUAGUCACAAACUGUCACAUGUGAUUGUGACGCAUUCGGCUGGUCGGUUGACCCCAGCAGUUGUCAUUUUGUGGCCAGAGU ......................................(((.((((((((.(((((((......)))))))....((((((.(.((.....)).).))))))....)))))))).))).. (-22.68 = -22.40 + -0.28)


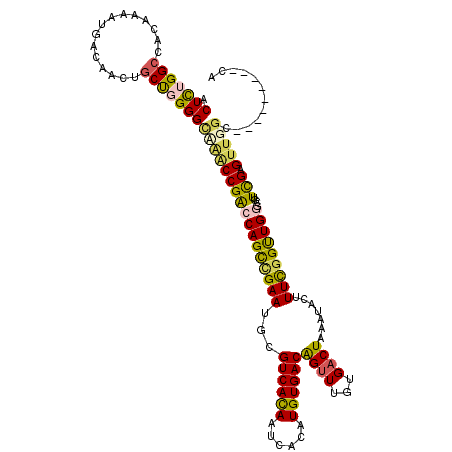
| Location | 21,947,193 – 21,947,304 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.74 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21947193 111 - 23771897 ACUCUGGCCGCAAAAUGACAACCGCUGGGGUUAACCGACCAGCCGAAUGCGUCACAAUCACAUGUGACAGUUUGUGACUAAAUACUUUCGGUUGGUCGUUCGAGUUGGC---------UA ....(((((((........((((.....))))...((((((((((((...((((((......))))))((((...)))).......)))))))))))).....).))))---------)) ( -38.60) >DroSec_CAF1 68867 111 - 1 ACUCUGGCCGCAAAAUGACAACUGCUGGGGCCAACCGACCAGCCGAAUGCGUCACAAUCACAUGUGACAGUUUGUGACUAAAUACUUUCGGUUGGUCGUUCGAGUUGGC---------UA (((((((((.((.............)).)))))..((((((((((((...((((((......))))))((((...)))).......))))))))))))...))))....---------.. ( -42.12) >DroEre_CAF1 68648 119 - 1 ACUUUGGCCACAAAAUGACAACUGCU-GGGUAGACCGACCAGCUGAAUGCGUCAUAAUCACAUGUGACAGUUUGCGACUAAAUACUUUCCGUUGGGCGUUCGAGUUGGCCAUUGUGGCCA ....((((((((..(((.((((((((-((.........)))))(((((((((((((......))))))......((((..((....))..)))).))))))))))))..))))))))))) ( -44.00) >DroYak_CAF1 84719 111 - 1 ACUCUGGGCACAAAAUGACAACUGCUCGGGCAGACCGACCAGCUGAAUGCGUCACAAUCACAUGUGACAGUUUGUGACUAAAUACUUUCGGUUGCGCUUUUGAGCUGGU---------CA ..((((((((............))))))))......((((((((.((.((((((((......)))))).....(..((..((....))..))..)))..)).)))))))---------). ( -40.60) >DroAna_CAF1 76983 110 - 1 ACUCUGGCCACAA-AUGACAACUGCCGGGGACAGCCGGCCAGUCGAAUGCGUCACAAUCACAUGUGACGGCUUGUGACUAAAUACUUUUGGCUGCAAAUUCGUGUCGGG---------CA ......(((....-.(((((((((((((......)))).))))(((((.(((((((......)))))))..((((..(((((....)))))..))))))))))))))))---------). ( -38.00) >consensus ACUCUGGCCACAAAAUGACAACUGCUGGGGCAAACCGACCAGCCGAAUGCGUCACAAUCACAUGUGACAGUUUGUGACUAAAUACUUUCGGUUGGGCGUUCGAGUUGGC_________CA .(((((((...............)))))))(((((((((((((((((...((((((......))))))((((...)))).......)))))))))....))).)))))............ (-24.42 = -24.74 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:27 2006