
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,922,345 – 21,922,457 |
| Length | 112 |
| Max. P | 0.767466 |

| Location | 21,922,345 – 21,922,440 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -15.47 |
| Energy contribution | -14.37 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21922345 95 + 23771897 GUU-UGUUUGUAUGUCGGUUUCAGUUUGCGGAAAUGGUUUUGCAUUUUGCAUUUGCUGCCAUUGCAACUGCAACUGCAACACCGAGCUGCAAGUUC ...-..((((((..((((((((((((.((((.((((((...(((.........))).))))))....))))))))).)).)))))..))))))... ( -29.60) >DroSec_CAF1 43159 95 + 1 GUU-UGUUUGUCUGUCGGUUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUUGCUGGCAUUGCAACUGCAACUGCAACACCGAGCUGCAAGUUC ...-..................(((((((((.....((.(((((..(((((.((((.......)))).))))).))))).))....))))))))). ( -27.80) >DroSim_CAF1 43481 95 + 1 GUU-UGUUUGUCUGUCGGUUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUUGCUGGCAUUGCAACUGCAACUGCAACACCGAGCUGCAAGUUC ...-..................(((((((((.....((.(((((..(((((.((((.......)))).))))).))))).))....))))))))). ( -27.80) >DroEre_CAF1 44183 94 + 1 GUU-UGUUUGUCUGUCGGCUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUG-CUGCCAUUGCAAUUGCAACUGCAACACCGAGCUGCAAGUUC ...-.....(((....)))...(((((((((.....((.(((((..((((((((-(.......)))).))))).))))).))....))))))))). ( -27.80) >DroYak_CAF1 51347 95 + 1 GUU-UGUCUGUCUGUCGGCUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUGGUUGCCAUCGCAACUGCGACUGCAAUAUCGAGCUGCAAGUUC ...-.....(((....)))...(((((((((...(((..(((((..(((((...(((((....)))))))))).)))))...))).))))))))). ( -29.10) >DroMoj_CAF1 61033 77 + 1 GUUCUGCUUGUUUGCCAUGCCCAUUUUGCGAGAAUUGUUUUGUAUUUUGCAACUGUU------------------AUAGAAGCGGGUGGCAAUU-U ...........(((((..((((...((((((((((......)).))))))))..(((------------------.....))))))))))))..-. ( -18.00) >consensus GUU_UGUUUGUCUGUCGGUUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUUGCUGCCAUUGCAACUGCAACUGCAACACCGAGCUGCAAGUUC ..........(((((.(((....))).)))))......((((((.((((...((((.....((((....))))..))))...)))).))))))... (-15.47 = -14.37 + -1.10)

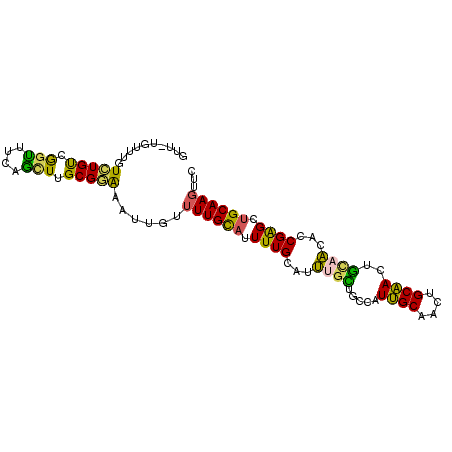

| Location | 21,922,345 – 21,922,440 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -11.24 |
| Energy contribution | -12.70 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21922345 95 - 23771897 GAACUUGCAGCUCGGUGUUGCAGUUGCAGUUGCAAUGGCAGCAAAUGCAAAAUGCAAAACCAUUUCCGCAAACUGAAACCGACAUACAAACA-AAC ....(((....(((((....(((((((.(((((....)))))...(((.....)))...........)).)))))..)))))....)))...-... ( -24.20) >DroSec_CAF1 43159 95 - 1 GAACUUGCAGCUCGGUGUUGCAGUUGCAGUUGCAAUGCCAGCAAAUGCAAAAUGCAAAACAAUUUCCGCAAGCUGAAACCGACAGACAAACA-AAC ...(((((.(((.((((((((((......)))))))))))))...(((.....)))...........)))))(((.......))).......-... ( -26.50) >DroSim_CAF1 43481 95 - 1 GAACUUGCAGCUCGGUGUUGCAGUUGCAGUUGCAAUGCCAGCAAAUGCAAAAUGCAAAACAAUUUCCGCAAGCUGAAACCGACAGACAAACA-AAC ...(((((.(((.((((((((((......)))))))))))))...(((.....)))...........)))))(((.......))).......-... ( -26.50) >DroEre_CAF1 44183 94 - 1 GAACUUGCAGCUCGGUGUUGCAGUUGCAAUUGCAAUGGCAG-CAAUGCAAAAUGCAAAACAAUUUCCGCAAGCUGAAGCCGACAGACAAACA-AAC ...(((.((((((((((((((.(((((....))))).))))-)).(((.....))).........)))..))))))))..............-... ( -27.10) >DroYak_CAF1 51347 95 - 1 GAACUUGCAGCUCGAUAUUGCAGUCGCAGUUGCGAUGGCAACCAAUGCAAAAUGCAAAACAAUUUCCGCAAGCUGAAGCCGACAGACAGACA-AAC ....((((((((((((......)))).))))))))((((......(((.....))).......((((....)..)))))))...........-... ( -22.40) >DroMoj_CAF1 61033 77 - 1 A-AAUUGCCACCCGCUUCUAU------------------AACAGUUGCAAAAUACAAAACAAUUCUCGCAAAAUGGGCAUGGCAAACAAGCAGAAC .-..((((((((((((.....------------------...)))(((...................)))....)))..))))))........... ( -12.11) >consensus GAACUUGCAGCUCGGUGUUGCAGUUGCAGUUGCAAUGGCAGCAAAUGCAAAAUGCAAAACAAUUUCCGCAAGCUGAAACCGACAGACAAACA_AAC ....(((....(((((....(((((((.((((......))))...(((.....)))...........)).)))))..)))))....)))....... (-11.24 = -12.70 + 1.46)

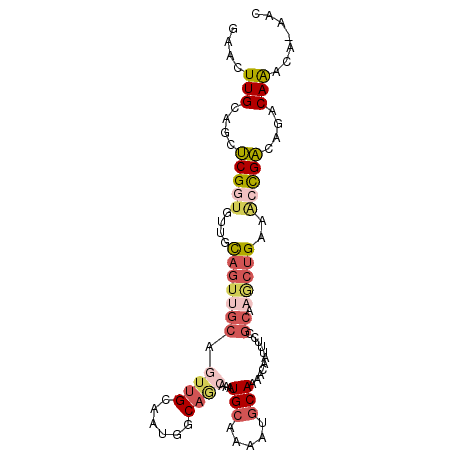

| Location | 21,922,362 – 21,922,457 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.47 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21922362 95 + 23771897 UUUCAGUUUGCGGAAAUGGUUUUGCAUUUUGCAUUUGCUGCCAUUGCAACUGCAACUGCAACACCGAGCUGCAAGUUCUGUGCAAGUUCUGUUUA ...((((((((((...((((.(((((..(((((.((((.......)))).))))).))))).))))..)))))))..)))............... ( -29.50) >DroSec_CAF1 43176 95 + 1 UUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUUGCUGGCAUUGCAACUGCAACUGCAACACCGAGCUGCAAGUUCUGUGUGCGCCCUGUUUA ....(((((((((.....((.(((((..(((((.((((.......)))).))))).))))).))....))))))))).................. ( -27.80) >DroSim_CAF1 43498 95 + 1 UUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUUGCUGGCAUUGCAACUGCAACUGCAACACCGAGCUGCAAGUUCUGUGUGCGCCCUGUUUA ....(((((((((.....((.(((((..(((((.((((.......)))).))))).))))).))....))))))))).................. ( -27.80) >DroEre_CAF1 44200 92 + 1 CUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUG-CUGCCAUUGCAAUUGCAACUGCAACACCGAGCUGCAAGUUCU--GUGCGCCCUGUUUA ....(((((((((.....((.(((((..((((((((-(.......)))).))))).))))).))....)))))))))..--.............. ( -26.80) >DroYak_CAF1 51364 95 + 1 CUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUGGUUGCCAUCGCAACUGCGACUGCAAUAUCGAGCUGCAAGUUCCGUGUGAGCCCUGUUUA .....((((((((((.....((((((.((((.....(((((..((((....))))..)))))..)))).))))))))))))..))))........ ( -28.30) >DroAna_CAF1 50578 87 + 1 UCUUAGUUAGUUGAAAUU-UGCUGGAU-UUGCUGUAGCUGUUGUUGCAACAGAAAGCGGAA-----AGUGGCAAGGGCG-GGGGCGGAUUGUUUA .......((((.......-.))))(((-(((((.(.(((.((((..(..............-----.)..)))).))).-).))))))))..... ( -22.16) >consensus UUUCAGCUUGCGGAAAUUGUUUUGCAUUUUGCAUUUGCUGCCAUUGCAACUGCAACUGCAACACCGAGCUGCAAGUUCUGUGUGCGCCCUGUUUA ....(((((((((.....((.(((((..(((((...((.......))...))))).))))).))....))))))))).................. (-16.20 = -16.47 + 0.27)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:13:19 2006